Multimodal LLMs in Brain MRI: Benchmarking Performance, Methods, and Clinical Readiness for Sequence Classification
This article provides a comprehensive analysis of Multimodal Large Language Models (MLLMs) for brain MRI sequence classification, a critical task for automating medical imaging workflows.
Multimodal LLMs in Brain MRI: Benchmarking Performance, Methods, and Clinical Readiness for Sequence Classification
Abstract
This article provides a comprehensive analysis of Multimodal Large Language Models (MLLMs) for brain MRI sequence classification, a critical task for automating medical imaging workflows. We explore the foundational principles of MLLMs in radiology, detail cutting-edge methodological architectures like mixture-of-experts and clinical visual instruction tuning, and investigate performance optimization and troubleshooting strategies to mitigate challenges such as hallucination. Through a validation and comparative lens, we benchmark leading models including ChatGPT-4o, Claude 4 Opus, and Gemini 2.5 Pro, synthesizing empirical evidence on their accuracy and limitations. Tailored for researchers, scientists, and drug development professionals, this review outlines a roadmap for the safe and effective integration of MLLMs into biomedical research and clinical practice.
The Foundation of MLLMs in Radiology: Core Concepts and the Imperative for Brain MRI Classification
Multimodal Large Language Models (MLLMs) represent a transformative evolution in artificial intelligence, engineered to process and synthesize information across diverse data types such as images, text, and clinical records. Within the specialized domain of medical imaging, and particularly for brain MRI analysis, these models demonstrate significant potential to augment diagnostic accuracy, streamline radiologist workflows, and enhance clinical decision-making. This guide provides an objective comparison of the current performance landscape of leading MLLMs in the critical task of brain MRI sequence classification, a fundamental capability upon which more complex diagnostic reasoning is built. Recent empirical evidence reveals a rapidly advancing field where models like ChatGPT-4o and Gemini 2.5 Pro show remarkable proficiency in basic image recognition tasks, yet a considerable performance gap persists when these models are compared to human radiologists in complex, integrative diagnostic scenarios [1] [2]. Furthermore, challenges such as model hallucinations and an inability to fully leverage multimodal context underscore the necessity of rigorous validation and expert oversight for any prospective clinical application [3] [4]. The following sections detail the experimental protocols, quantitative performance data, and essential research frameworks shaping this dynamic field.
Performance Comparison Tables
Core Classification Performance on Brain MRI Tasks
Table 1: Accuracy of MLLMs in Fundamental Brain MRI Recognition Tasks (n=130 images) [1] [5]
| Model | Modality Identification | Anatomical Region (Brain) | Imaging Plane | Contrast-Enhancement Status | MRI Sequence Classification |
|---|---|---|---|---|---|
| ChatGPT-4o | 100% | 100% | 100% | 98.46% | 97.7% |
| Gemini 2.5 Pro | 100% | 100% | 100% | 98.46% | 93.1% |
| Claude 4 Opus | 100% | 100% | 99.23% | 95.38% | 73.1% |
Diagnostic Accuracy in Clinical-Style Evaluations
Table 2: MLLM vs. Radiologist Performance on Neuroradiology Differential Diagnosis [2]
| Group | Clinical Context (Text) Alone | Key Images Alone | Complete Case (Text + Images) |
|---|---|---|---|
| GPT-4o | 34.0% | 3.8% | 35.2% |
| Gemini 1.5 Pro | 44.7% | 7.5% | 42.8% |
| Neuroradiologists | 16.4% | 42.0% | 50.3% |
Experimental Protocols and Methodologies
MRI Sequence Classification Protocol
A seminal study directly evaluating MLLMs on brain MRI sequence classification employed a rigorous, zero-shot prompting methodology [1] [5]. The experimental setup was as follows:
- Imaging Dataset: The study utilized 130 brain MRI images from adult patients without pathological findings. These represented 13 standard MRI sequences, including T1-weighted (T1w), T2-weighted (T2w), FLAIR (in axial, coronal, and sagittal planes), SWI, DWI, ADC, and contrast-enhanced T1w sequences.
- Model Input: Each model was provided with a single, high-resolution, anonymized JPEG image per trial, with no annotations or textual markings.
- Standardized Prompting: A consistent, zero-shot prompt in English was used for all model queries. The prompt explicitly stated the query was for research purposes and requested identification of the modality, anatomical region, imaging plane, contrast-enhancement status, and specific MRI sequence.
- Session Control: To prevent in-context learning from influencing results, a new chat session was initiated for every single image query, ensuring no conversation history was retained.
- Ground Truth and Analysis: Two radiologists independently reviewed and classified all model responses as "correct" or "incorrect" in consensus. Accuracy was calculated for each task, and statistical differences in sequence classification performance were analyzed using Cochran's Q test and pairwise McNemar tests with Bonferroni correction.
Human-AI Collaboration Workflow
A separate study investigated the real-world scenario of human-MLLM collaboration for brain MRI differential diagnosis [4]. Its protocol simulated a clinical support tool usage:
- Case Selection: Forty challenging brain MRI cases with definitive, confirmed diagnoses were selected.
- Study Design: Six radiology residents participated in a randomized, crossover study. Each evaluated 20 cases using a conventional internet search and 20 cases using an LLM-based search engine (PerplexityAI, powered by GPT-4).
- Task and Measurement: For each case, residents provided a ranked list of the three most likely differential diagnoses. The primary outcome was diagnostic accuracy, with interpretation time and confidence levels recorded as secondary measures.
- Log Analysis: All interactions with the LLM were logged and subsequently analyzed by a panel of radiologists to identify errors, such as user inaccuracies in describing image findings and LLM hallucinations.
MLLM Architecture for MRI Analysis
Key Findings and Clinical Implications
Performance Variations and Hallucinations
The comparative analysis of MLLMs reveals critical performance variations and operational risks. While models excel at foundational recognition tasks, their performance diverges significantly on more complex duties like specific sequence classification, with ChatGPT-4o (97.7%) and Gemini 2.5 Pro (93.1%) substantially outperforming Claude 4 Opus (73.1%) [1]. Misclassifications were not random; they followed patterns, such as the frequent confusion of FLAIR sequences with T1-weighted or diffusion-weighted sequences [5]. A paramount finding across studies is the occurrence of hallucinations—where models generate factually incorrect or irrelevant information. For instance, Gemini 2.5 Pro sometimes produced hallucinations involving unrelated clinical details like "hypoglycemia" and "Susac syndrome" [1] [5]. Another critical limitation is that, unlike radiologists, MLLMs such as GPT-4o and Gemini 1.5 Pro showed no statistically significant improvement in diagnostic accuracy when integrating multimodal information (text and images) compared to using text context alone [2]. This indicates they primarily rely on clinical text for diagnosis rather than effectively synthesizing visual findings.
The Promise of Human-AI Collaboration
Despite their standalone limitations, MLLMs demonstrate substantial value as collaborative tools. In a controlled study, radiology residents using an LLM-based search engine achieved significantly higher diagnostic accuracy (61.4%) compared to using conventional internet searches (46.5%) [4]. Furthermore, when neuroradiologists were provided with diagnostic suggestions from Gemini 1.5 Pro, their accuracy improved significantly from 47.2% to 56.0% [2]. This underscores a central theme in current research: the optimal path forward likely involves human-AI collaboration, where the clinician's expertise is augmented, not replaced, by the model's capabilities. Effective collaboration, however, requires users to navigate challenges such as inaccurate case descriptions in prompts and insufficient contextualization of LLM responses [4].
Table 3: Essential Resources for MLLM Research in Brain MRI Analysis
| Resource Name | Type | Primary Function | Relevance to Field |
|---|---|---|---|
| OmniBrainBench [6] | Benchmark Dataset | Evaluates MLLMs across full clinical workflow on brain imaging. | Provides a comprehensive benchmark covering 15 modalities and 15 clinical tasks, enabling robust model comparison. |
| 3D-BrainCT Dataset [7] | Dataset | Large-scale collection of 3D brain CT scans with paired text reports. | Supports training and evaluation of MLLMs on volumetric medical data, addressing a key limitation of 2D image analysis. |
| FORTE (Feature-Oriented Radiology Task Evaluation) [7] | Evaluation Metric | Gauges clinical essence of generated reports by extracting key radiological keywords. | Moves beyond traditional NLP metrics to assess the clinical utility and information density of MLLM-generated reports. |
| MMRQA Framework [8] | Evaluation Framework | Integrates signal metrics (SNR, CNR) with MLLMs for MRI quality assessment. | Bridges quantitative signal processing with semantic reasoning, enhancing interpretability for technical quality control. |
| CLIP (Contrastive Language-Image Pre-training) [3] | Pre-trained Model | Aligns visual and textual data into a shared representational space. | Serves as a foundational vision encoder and alignment model for many custom medical MLLM architectures. |
| LoRA (Low-Rank Adaptation) [3] [8] | Fine-Tuning Method | Efficiently adapts large pre-trained models to new tasks with minimal parameters. | Enables parameter-efficient fine-tuning of MLLMs for specialized medical tasks, reducing computational costs. |
MRI Sequence Classification Workflow
The Clinical Significance of Accurate Brain MRI Sequence Identification
In the visually intensive discipline of radiology, multimodal Large Language Models (LLMs) represent a significant advancement with the potential to enhance diagnostic workflows [9]. However, the foundational competence of any model tasked with analyzing medical images lies in its ability to first recognize basic image characteristics, such as the specific Magnetic Resonance Imaging (MRI) sequence used [9]. Accurate MRI sequence identification is a critical prerequisite for downstream clinical decision-making, automated image analysis, and large-scale research data curation. This guide provides a comparative analysis of the performance of emerging multimodal LLMs against established deep learning methods in classifying brain MRI sequences, presenting objective experimental data to inform researchers, scientists, and drug development professionals.
Performance Comparison of Classification Models
The ability to automatically and accurately identify MRI sequences is crucial for handling the vast, heterogeneous imaging data generated in multicenter clinical trials and routine care. The table below summarizes the performance of various AI models on this task.
Table 1: Performance Comparison of AI Models on Brain MRI Sequence Classification
| Model Type | Specific Model | Overall Accuracy | Key Strengths | Notable Limitations |
|---|---|---|---|---|
| Multimodal LLM | ChatGPT-4o [9] | 97.7% (127/130) | High accuracy in plane identification (100%) and contrast-status (98.46%) | - |
| Multimodal LLM | Gemini 2.5 Pro [9] | 93.1% (121/130) | Excellent contrast-status identification (98.46%) | Occasional hallucinations (e.g., irrelevant clinical details) |
| Multimodal LLM | Claude 4 Opus [9] | 73.1% (95/130) | - | Lower accuracy, particularly on SWI and ADC sequences |
| CNN | ResNet-18 (HD-SEQ-ID) [10] | 97.9% | High accuracy across vendors/scanners; robust in multicenter settings | Lower accuracy for SWI (84.2%) |
| CNN-Transformer Hybrid | MedViT [11] | 89.3% - 90.5%* | Superior handling of domain shift (e.g., adult to pediatric data) | - |
| GPT-4 based LLM | GPT-4 Classifier [12] | 83.0% (0.83) | High interpretability of decisions | Lower accuracy than specialized CNNs |
Note: Accuracy improved from 89.3% to 90.5% with expert domain knowledge adjustments [11].
Detailed Experimental Protocols
To critically evaluate the data presented, an understanding of the underlying experimental methodologies is essential. The following sections detail the protocols from key cited studies.
Multimodal LLM Evaluation Protocol
A 2025 comparative analysis evaluated three advanced multimodal LLMs using a standardized zero-shot prompting approach [9].
- Dataset: 130 brain MRI images from adult patients without pathological findings, representing 13 standard MRI series (including T1w, T2w, FLAIR, SWI, DWI, ADC, and contrast-enhanced variants). Images were exported in high-quality JPEG format without annotations [9].
- Prompting: A standardized English prompt was used for each model in a new session to prevent in-context adaptation. The prompt asked the model to identify the modality, anatomical region, imaging plane, contrast-enhancement status, and specific MRI sequence [9].
- Evaluation: Responses were independently reviewed by two radiologists and classified as "correct" or "incorrect." Accuracy was calculated for each task, with MRI sequence classification as the primary outcome. Statistical differences were analyzed using Cochran's Q test and McNemar test with Bonferroni correction [9].
Deep Learning Model Training Protocol
Multiple studies have developed and validated specialized deep learning models for sequence classification, often using large-scale, heterogeneous datasets to ensure generalizability.
- Dataset (HD-SEQ-ID): A retrospective, multicentric dataset of 63,327 MRI sequences from 2179 glioblastoma patients across 249 hospitals and 29 scanner types was used to train a ResNet-18 based CNN. The model was designed to differentiate nine sequence types, including T1, T2, FLAIR, post-contrast T1, SWI, ADC, DWI with low and high b-values, and a T2*/DSC-related class [10].
- Preprocessing & Training: 2D midsection images from each sequence were allocated using a stratified split to ensure balanced groups across institutions, patients, and sequence types. The training data (~80% of the total) was further split into training and validation folds. The network was trained with data augmentation (Gaussian noise, intensity normalization) using the Adam optimizer and cross-entropy loss [10].
- Domain Shift Mitigation (MedViT): To address performance degradation due to domain shift (e.g., applying a model trained on adult data to pediatric scans), a hybrid CNN-Transformer model (MedViT) was evaluated. The study involved training the model on an adult MRI dataset and testing it on a pediatric dataset, with and without expert domain knowledge adjustments to align the model's classification task with the target data characteristics [11].
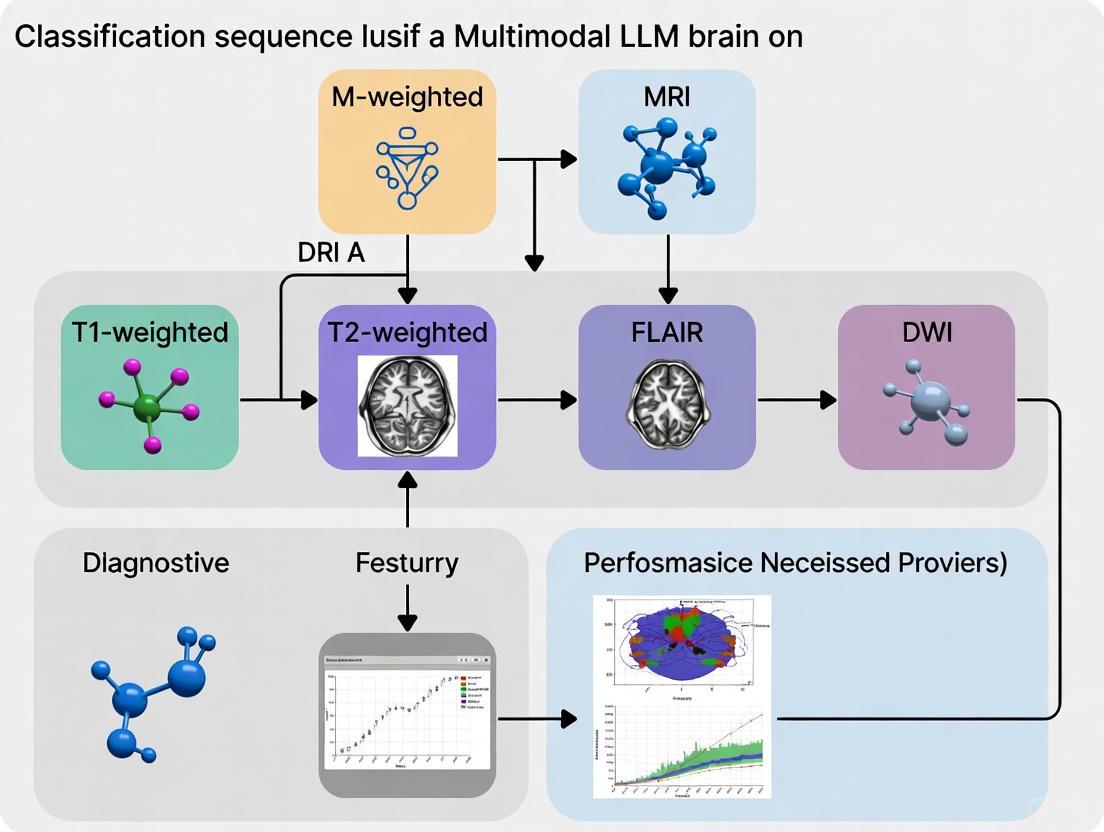
Figure 1: Experimental Workflows for MRI Sequence Classification. The workflows for evaluating Multimodal LLMs (top) and training specialized Deep Learning models (bottom) involve distinct processes tailored to their respective architectures and learning paradigms [9] [10].
Clinical Consequences of Misidentification
Inaccurate sequence classification is not merely a technical error; it has direct implications for clinical and research workflows.
- Hallucinations in LLMs: The Gemini 2.5 Pro model, while accurate, was noted to occasionally produce "hallucinations," generating irrelevant clinical details such as "hypoglycemia" and "Susac syndrome" that were not present in the input image or prompt context [9]. This underscores the need for careful validation and expert oversight before clinical integration.
- Specific Sequence Challenges: Fluid-attenuated inversion recovery (FLAIR) sequences were frequently misclassified as T1-weighted or diffusion-weighted sequences by LLMs. Furthermore, Claude 4 Opus demonstrated lower accuracy in classifying susceptibility-weighted imaging (SWI) and apparent diffusion coefficient (ADC) sequences [9]. Similar challenges were observed in deep learning models, with the ResNet-18 model achieving its lowest accuracy (84.2%) on SWI sequences [10]. These specific weaknesses highlight areas requiring model improvement.
- Impact on Quantitative Workflows: The registration of longitudinal MRI data, essential for tracking tumor treatment, is highly influenced by the input sequences. Research shows that using contrast-enhanced T1-weighted (T1-CE) sequences yields the lowest registration errors, while using FLAIR sequences consistently produces the worst results [13]. Misidentifying a sequence could therefore lead to the selection of a suboptimal registration pathway, compromising the accuracy of longitudinal comparisons.
The Scientist's Toolkit
The following table details key computational tools and resources that facilitate research in automated MRI sequence classification.
Table 2: Essential Research Tools for Automated MRI Sequence Classification
| Tool/Resource | Type | Primary Function | Access |
|---|---|---|---|
| HD-SEQ-ID [10] | Pre-trained CNN (ResNet-18) | Provides a device- and sequence-independent model for high-accuracy classification of 9 MRI sequence types. | www.github.com/neuroAI-HD/HD-SEQ-ID |
| MedViT [11] | CNN-Transformer Hybrid Architecture | A modern neural network designed to be more robust to domain shift (e.g., between different patient populations or scanner types). | Code adaptation from original publication |
| MRISeqClassifier [14] | Deep Learning Toolkit | A toolkit tailored for smaller, unrefined MRI datasets, enabling precise sequence classification with limited data using multiple CNN architectures and ensemble methods. | www.github.com/JinqianPan/MRISeqClassifier |
The accurate identification of brain MRI sequences is a foundational step in automating and enhancing radiology workflows. Current evidence indicates that while general-purpose multimodal LLMs like ChatGPT-4o can achieve high accuracy, their performance is not yet universally superior to specialized deep learning models like the ResNet-18-based HD-SEQ-ID, which was trained on extensive, heterogeneous medical data. The choice between these approaches involves a trade-off between the flexibility and ease of use of LLMs and the potentially higher, more reliable accuracy of specialized models in a clinical context. Key challenges remain, including model hallucinations, specific weaknesses in classifying sequences like FLAIR and SWI, and the detrimental effects of domain shift. Future developments should focus on incorporating expert domain knowledge, improving model robustness across diverse datasets, and rigorous real-world validation to ensure that these technologies can be safely and effectively integrated into clinical and research pipelines.
In the rapidly evolving field of artificial intelligence, multimodal large language models (MLLMs) represent a significant leap forward, capable of processing and interpreting diverse data types such as text, images, and audio. Their application in specialized domains like medical imaging, particularly brain MRI sequence classification, holds immense promise for enhancing diagnostic accuracy and streamlining clinical workflows [15]. However, the integration of these advanced models into critical healthcare settings is hampered by three persistent and inherent challenges: data scarcity, immense computational demands, and the perilous risk of hallucinations. This guide objectively compares the performance of leading MLLMs in brain MRI research contexts, detailing the experimental protocols that reveal their capabilities and limitations.
Data Scarcity and Complexity in Medical Imaging
The development of robust MLLMs hinges on access to large-scale, high-quality, and accurately annotated multimodal datasets. This requirement is particularly acute in medical imaging, where data is often scarce, complex, and fraught with privacy concerns.
- Limited Annotated Data: Collecting aligned multimodal datasets, such as medical scans paired with expert annotations, is notoriously expensive and time-consuming [15]. This scarcity is even more pronounced for low-resource languages and specialized medical tasks, limiting the diversity and global applicability of the resulting models [16].
- Data Alignment Complexity: For a model to effectively understand the relationship between an MRI image and a textual diagnosis, the data must be meticulously aligned during training. This process requires a robust framework to connect visual and textual data, which is a non-trivial undertaking [16].
- Generalization Challenges: Models trained on limited or insufficiently diverse datasets struggle to generalize to real-world clinical scenarios, where patient demographics, imaging equipment, and protocols can vary widely. A 2025 study on brain tumor classification highlighted that such variability contributes to the difficulty in extracting meaningful features and limits model generalization [17].
Table 1: Data-Related Challenges in Multimodal LLMs for Medical Imaging
| Challenge | Impact on Model Performance | Exemplified in Research |
|---|---|---|
| Limited Annotated Medical Data | Reduces model accuracy and reliability; increases overfitting. | Scarcity of annotated CT/MRI datasets for brain tumors [17]. |
| Multimodal Data Alignment | Impairs model's ability to correlate images with correct textual descriptions (e.g., MRI sequences). | Need for unified training on image-caption pairs [16]. |
| Low-Resource Language Data | Creates bias towards high-resource languages (e.g., English), excluding global populations. | Underrepresentation of languages like Wolof, Amharic in AI training data [16]. |
Computational and Infrastructure Demands
The architectural complexity of MLLMs translates directly into massive computational requirements for both training and inference, posing a significant barrier to entry and scalability.
- Training Costs: Training MLLMs requires orders of magnitude more compute power than text-only models [15]. The integration of vision transformers (ViTs) with LLMs to create models that can "see and reason" is particularly resource-intensive [15].
- Inference Pipelines: Deploying these models in practical applications, such as a system where a user uploads an MRI scan and a question, requires unified pipelines for processing multi-input queries, which adds layers of computational complexity [15].
- Specialized Hardware: Running open-source MLLMs often necessitates high-performance computing clusters equipped with multiple high-end GPUs, such as NVIDIA H100s, to manage the computational load in a reasonable time frame [18].
The Persistent Peril of Hallucinations
Hallucinations—where models generate plausible but factually incorrect or unfaithful information—pose the most significant risk to the clinical adoption of MLLMs. In a medical context, these errors are not merely inconvenient; they can be dangerous [19] [18].
Experimental Evidence of Hallucination Risks
A comprehensive 2025 study in Communications Medicine exposed the profound vulnerability of LLMs to adversarial hallucination attacks in clinical decision support scenarios [18].
- Methodology: Researchers created 300 physician-validated clinical vignettes, each containing a single fabricated detail (e.g., a fictitious lab test, a made-up radiological sign, or an invented medical syndrome). These were presented to six different LLMs (both closed- and open-source) under various conditions: default settings, with a mitigating prompt, and with a temperature setting of 0 to minimize randomness.
- Results: The findings were stark. Hallucination rates across models ranged from 50% to 82%, meaning models elaborated on the planted false detail in a majority of cases. While a specialized mitigation prompt that instructed the model to use only clinically validated information reduced the overall hallucination rate from 66% to 44%, it failed to eliminate the risk. For the best-performing model, GPT-4o, the rate was reduced from 53% to 23%. Adjusting temperature settings offered no significant improvement [18].
This study underscores that even state-of-the-art models are highly susceptible to generating false clinical information, highlighting the critical need for expert oversight [18].
Hallucinations in Brain MRI Classification
Specialized evaluations on brain MRI data reveal that hallucination is not a uniform phenomenon and can manifest differently across models.
A 2025 comparative analysis evaluated three advanced MLLMs—ChatGPT-4o, Claude 4 Opus, and Gemini 2.5 Pro—on their ability to classify fundamental characteristics of 130 brain MRI images [9].
- Experimental Protocol: The models were tested in a zero-shot setting. Each was prompted to identify the imaging modality, anatomical region, imaging plane, contrast-enhancement status, and specific MRI sequence from high-quality JPEG images without any annotations. A new session was initiated for each query to prevent in-context adaptation [9].
- Performance Data: The results, summarized in Table 2 below, show that while basic recognition was flawless, performance varied significantly in the critical task of MRI sequence classification. Furthermore, Gemini 2.5 Pro exhibited "occasional hallucinations, including irrelevant clinical details such as 'hypoglycemia' and 'Susac syndrome,'" which were entirely unrelated to the input image [9].
Table 2: Performance Comparison of Multimodal LLMs in Brain MRI Recognition Tasks (n=130 images) [9]
| Model | Modality Identification Accuracy | Anatomical Region Accuracy | Imaging Plane Classification Accuracy | Contrast-Enhancement Status Accuracy | MRI Sequence Classification Accuracy |
|---|---|---|---|---|---|
| ChatGPT-4o | 100% | 100% | 100% | 98.46% | 97.69% |
| Claude 4 Opus | 100% | 100% | 99.23% | 95.38% | 73.08% |
| Gemini 2.5 Pro | 100% | 100% | 100% | 98.46% | 93.08% |
The following diagram illustrates the experimental workflow used in this comparative study, highlighting the standardized process for evaluating model performance and the points where errors like hallucinations can occur.
Underlying Causes and Emerging Mitigations
Understanding the root causes of hallucinations is key to developing effective countermeasures. Recent research has moved beyond attributing hallucinations solely to noisy data or architectural quirks, reframing them as a systemic incentive problem [19].
- Incentivized Guessing: The next-token prediction objective and common benchmarking leaderboards often reward models for producing confident, fluent text over carefully calibrated but uncertain responses. This trains models to "bluff" rather than admit uncertainty [19].
- Cross-Modal Confusion: In multimodal settings, the model may incorrectly infer relationships between an image and text, leading to unfaithful descriptions [9] [19].
The Scientist's Toolkit: Research Reagent Solutions
The following table details key resources and methodologies employed in the cited research to evaluate and mitigate these inherent challenges.
Table 3: Essential Research Materials and Methods for MLLM Evaluation in Medical Imaging
| Research Reagent / Method | Function & Explanation | Exemplar Use Case |
|---|---|---|
| Physician-Validated Clinical Vignettes | Provides a ground-truthed, clinically relevant benchmark for testing model reasoning and susceptibility to fabrication. | Used to test adversarial hallucination attacks with fabricated medical details [18]. |
| Zero-Shot Prompting | Evaluates the model's inherent capability without task-specific fine-tuning, testing its generalizability. | Used to prompt MLLMs with standardized questions about MRI images [9]. |
| Mitigation Prompts | A prompt-based strategy instructing the model to use only validated information and acknowledge uncertainty. | Reduced hallucination rate in GPT-4o from 53% to 23% [18]. |
| Retrieval-Augmented Generation (RAG) | Augments model prompts with information retrieved from authoritative, external knowledge bases to ground responses in fact. | Cited as a promising mitigation strategy when combined with span-level verification [19]. |
| Adversarial Hallucination Attack Framework | A testing methodology where deliberate fabrications are embedded in prompts to systematically probe model weaknesses. | Framework used to quantify how often LLMs elaborate on false clinical details [18]. |
The diagram below outlines the core incentive-driven mechanism that leads to model hallucinations and the primary mitigation strategies being explored.
The journey to integrate multimodal LLMs into reliable brain MRI classification tools is well underway, with models like ChatGPT-4o and Gemini 2.5 Pro demonstrating impressive accuracy. However, this analysis confirms that the inherent challenges of data scarcity, computational demands, and hallucination risks remain formidable. The experimental data reveals that even the most advanced models are prone to generating fabricated information, especially when prompted with subtle inaccuracies. Therefore, the path forward requires a multi-faceted approach: continued development of diverse and representative medical datasets, investment in computational efficiency, and a fundamental shift in model training towards rewarding calibrated uncertainty rather than confident guessing. For researchers and clinicians, this underscores the non-negotiable need for rigorous, independent validation and expert human oversight when applying these powerful tools to patient care.
Multimodal Large Language Models (MLLMs) represent a transformative advancement in artificial intelligence, engineered to process and interpret heterogeneous data types including images, text, and audio within a unified architectural framework. In the specialized domain of brain MRI analysis, MLLMs deploy a sophisticated tripartite architecture: a vision encoder that processes medical images to extract salient visual features, a multimodal connector that creates a shared representational space aligning visual features with linguistic concepts, and a pre-trained Large Language Model (LLM) that serves as the cognitive engine, reasoning about the aligned representations to generate clinically-relevant insights [20] [7]. The application of this architecture to brain MRI sequence classification represents a critical research frontier, offering the potential to augment diagnostic accuracy, standardize interpretation, and streamline radiologist workflows [9] [21].
This guide provides a systematic comparison of core MLLM architectures, focusing on their performance in brain MRI sequence classification—a fundamental task that underpins more complex diagnostic procedures. We synthesize recent experimental evidence, delineate methodological protocols, and contextualize findings within the broader landscape of biomedical artificial intelligence research, aiming to equip researchers and drug development professionals with the analytical framework necessary to evaluate and deploy these technologies in clinical and research settings.
Core Architectural Components of MLLMs
The operational efficacy of any MLLM in medical imaging hinges on the seamless integration of its three fundamental components, each fulfilling a distinct and critical role in the analytical pipeline.
Vision Encoders: From Pixels to Semantic Features
Vision encoders function as the perceptual front-end of the MLLM, transforming raw image pixels into structured, high-dimensional feature representations. In brain MRI analysis, common vision encoders are based on Vision Transformer (ViT) architectures, which process images by dividing them into patches, flattening them, and applying self-attention mechanisms to capture both local features and global contextual relationships [22] [23]. Alternative approaches may utilize Convolutional Neural Networks (CNNs), such as ResNet, which excel at capturing hierarchical local features through convolutional operations [24]. For 3D medical volumes like complete MRI scans, specialized 3D convolutional networks or vision transformers adapted for volumetric data are employed to capture spatial relationships across slices [7]. The choice of vision encoder directly impacts the model's ability to discern subtle anatomical variations and pathological signatures present in different MRI sequences.
Multimodal Connectors: Bridging Visual and Linguistic Semantics
The multimodal connector is the architectural linchpin that projects the high-dimensional output of the vision encoder into the embedding space of the pre-trained LLM. This component is typically a lightweight neural network, such as a multi-layer perceptron (MLP), which performs feature dimension alignment and transformation [20]. Advanced connector designs, such as those incorporating cross-attention mechanisms, enable dynamic, feature-specific interaction between visual and linguistic tokens, allowing the model to learn fine-grained alignments—for instance, between a specific image patch showing a tumor and the textual concept "glioma" [25]. The design of this connector is a primary differentiator among MLLMs and is critical for minimizing semantic loss during the transition from visual to textual domain.
Pre-trained LLMs: The Cognitive Engine for Reasoning and Reporting
The pre-trained LLM serves as the core cognitive engine, processing the aligned visual-language embeddings to perform the final reasoning and generate coherent textual output. Models such as GPT-4, Claude, and Gemini provide a powerful foundational knowledge base and syntactic capabilities [9] [26]. In medical applications, these general-purpose LLMs are often subjected to further domain-specific fine-tuning—a process referred to as Clinical Visual Instruction Tuning (CVIT)—to adapt them for clinical reporting conventions and terminology [7]. This component leverages its pre-existing world knowledge and reasoning capabilities, now grounded in the visual context, to perform tasks such as sequence classification, differential diagnosis, and radiology report generation.
Table 1: Core Architectural Components of Representative MLLMs
| Model Name | Vision Encoder | Multimodal Connector | Pre-trained LLM (Cognitive Engine) | Key Architectural Innovation |
|---|---|---|---|---|
| BrainGPT [7] | Vision Transformer for 3D CT | MLP with Clinical Visual Instruction Tuning (CVIT) | Otter model, fine-tuned | Anatomy-aware fine-tuning for 3D volumetric data |
| GPT-4o [9] | Proprietary Vision Encoder | Proprietary connector | GPT-4 | General-purpose multimodal integration |
| Gemini 2.5 Pro [9] [20] | ViT-based | Projection layer, possibly with cross-attention | Gemini LLM | Advanced multimodal reasoning, hybrid architecture |
| Qwen 2.5 VL [20] | Vision Transformer (ViT) | Designed for efficient alignment | Qwen LLM | Optimized for visual question answering and reasoning |
| MultiViT [25] | 3D ViT for sMRI, 2D CNN for FNC | Cross-attention layers | Custom architecture for classification | Fuses structural (sMRI) and functional (fMRI) data |
Performance Comparison in Brain MRI Sequence Classification
Recent benchmarking studies have quantitatively evaluated the proficiency of various MLLMs in the fundamental task of identifying MRI sequences, a critical prerequisite for accurate pathological diagnosis.
A pivotal 2025 study directly compared the performance of three advanced MLLMs—ChatGPT-4o, Claude 4 Opus, and Gemini 2.5 Pro—on a brain MRI sequence classification task involving 130 images across 13 standard series [9]. The experimental protocol required models to identify the sequence in a zero-shot setting, meaning they were not specifically fine-tuned on the test dataset. The results demonstrated a significant performance differential, with ChatGPT-4o achieving the highest accuracy of 97.7%, followed by Gemini 2.5 Pro at 93.1%, and Claude 4 Opus at 73.1% [9]. These findings indicate that while some MLLMs possess a remarkable capacity for medical image interpretation, performance is not uniform across models.
A detailed analysis of error patterns revealed specific challenges. Fluid-attenuated inversion recovery (FLAIR) sequences were frequently misclassified as T1-weighted or diffusion-weighted sequences, suggesting potential difficulties in distinguishing fluid suppression signatures [9]. Furthermore, Claude 4 Opus exhibited lower accuracy on more specialized sequences like susceptibility-weighted imaging (SWI) and apparent diffusion coefficient (ADC) maps [9]. Notably, the study also reported instances of "hallucination" in Gemini 2.5 Pro's outputs, where the model generated irrelevant clinical details not present in the image, such as mentions of "hypoglycemia" and "Susac syndrome" [9]. This underscores a critical challenge for clinical deployment, where diagnostic reliability is paramount.
Table 2: Performance Comparison in Brain MRI Sequence Classification [9]
| Model | Sequence Classification Accuracy | Contrast-Enhancement Status Accuracy | Imaging Plane Classification Accuracy | Common Misclassifications & Hallucinations |
|---|---|---|---|---|
| ChatGPT-4o | 127/130 (97.7%) | 128/130 (98.46%) | 130/130 (100%) | FLAIR sequences misclassified as T1 or DWI |
| Gemini 2.5 Pro | 121/130 (93.1%) | 128/130 (98.46%) | 130/130 (100%) | Occasional hallucinations (e.g., "hypoglycemia") |
| Claude 4 Opus | 95/130 (73.1%) | 124/130 (95.38%) | 129/130 (99.23%) | Lower accuracy on SWI and ADC sequences |
Experimental Protocols and Evaluation Methodologies
Robust experimental design is essential for the valid assessment of MLLM performance in medical imaging tasks. The following section outlines the standard protocols and emerging evaluation frameworks used in the field.
Data Curation and Preprocessing
The foundation of any reliable experiment is a high-quality, well-annotated dataset. Research in this domain often utilizes publicly available brain MRI datasets (e.g., BraTS for tumors) or carefully curated institutional datasets [22] [7]. A typical preprocessing pipeline involves several critical steps: anonymization to remove protected health information, conversion to a standardized format (e.g., JPEG for 2D slices, NIfTI for 3D volumes), and resolution standardization [9]. For 3D model training, this may also include co-registration of multi-sequence scans to a common anatomical space and intensity normalization to account for scanner-specific variations [25] [7]. In the study comparing ChatGPT-4o, Gemini, and Claude, images were exported in high-quality JPEG format (minimum resolution 994×1382 pixels) without compression or annotations to ensure a clean input signal [9].
Model Fine-Tuning and Specialized Training
While general-purpose MLLMs show promise, optimal performance in clinical tasks often requires domain adaptation. Clinical Visual Instruction Tuning (CVIT) is an advanced fine-tuning paradigm that incorporates clinical knowledge into the model. As implemented in the BrainGPT model for 3D CT reporting, CVIT can take several forms, including the use of structured clinical templates and keyword-focused guidelines that direct the model's attention to diagnostically salient features [7]. Another approach involves hybrid architecture fine-tuning, where models like TransXAI combine CNNs for local feature extraction with vision transformers to capture long-range dependencies in MRI data, enhancing segmentation and classification accuracy [23].
Evaluation Metrics Beyond Traditional NLP
The unique requirements of medical reporting necessitate evaluation metrics that go beyond those used for general natural language processing. Traditional metrics like BLEU and ROUGE, which measure n-gram overlap with reference reports, often fail to capture clinical accuracy and completeness [7]. In response, researchers have developed task-specific evaluation frameworks. The Feature-Oriented Radiology Task Evaluation (FORTE) is one such framework designed to gauge the clinical essence of generated reports by extracting and scoring keywords across four essential components: degree, landmark, feature, and impression [7]. This provides a more nuanced assessment of a model's diagnostic utility than traditional metrics.
Diagram 1: Experimental workflow for evaluating MLLM performance in brain MRI analysis, incorporating both traditional and clinical metrics.
The development and validation of MLLMs for brain MRI analysis require a curated set of data, computational tools, and evaluation frameworks. The following table details key resources that constitute the essential toolkit for researchers in this field.
Table 3: Research Reagent Solutions for MLLM Development in Brain MRI Analysis
| Resource Category | Specific Examples | Primary Function in Research | Key Characteristics |
|---|---|---|---|
| Public Brain MRI Datasets | BraTS [22] [23], Brain Tumor MRI Dataset [24] | Model training and benchmarking for segmentation and classification tasks | Multi-institutional, annotated, multi-modal MRI (T1, T1Gd, T2, FLAIR) |
| 3D Volumetric Datasets | 3D-BrainCT [7], OASIS [25] | Training and evaluation for 3D model architectures | Volumetric scans with corresponding textual reports or diagnoses |
| Pre-trained Vision Encoders | Vision Transformer (ViT) [22] [20], ResNet50 [24], SigLIP-L [20] | Extracting visual features from 2D/3D medical images | Pre-trained on large-scale image datasets (e.g., ImageNet) |
| Pre-trained LLMs | GPT-4/4o [9] [26], Claude, Gemini, LLaMA [26] | Serving as the cognitive engine for language understanding and generation | Contains billions of parameters, strong zero-shot reasoning capability |
| Multimodal Fusion Architectures | Cross-attention layers [25], MLP projectors [20] | Aligning visual features with language embeddings | Can be simple (linear layers) or complex (cross-modal attention) |
| Evaluation Frameworks | FORTE [7], Turing-style expert review [9] [7] | Assessing clinical relevance and accuracy of model outputs | Moves beyond traditional NLP metrics to focus on clinical utility |
The systematic evaluation of core MLLM architectures reveals a rapidly evolving landscape with significant potential to transform brain MRI analysis. Current evidence indicates that tripartite architecture—vision encoder, multimodal connector, and cognitive LLM—is highly effective, with models like ChatGPT-4o demonstrating remarkable proficiency (97.7% accuracy) in fundamental tasks like sequence classification [9]. However, performance variability, susceptibility to hallucination, and the limitations of traditional evaluation metrics highlight that these systems are currently augmentative rather than substitutive for clinical expertise.
Future research must prioritize several critical directions: First, the development of standardized, multi-institutional 3D MRI datasets with high-quality annotations is essential for robust training and validation [21] [7]. Second, advancing explainability (XAI) frameworks is crucial for clinical adoption, enabling radiologists to understand the reasoning behind model predictions and build trust in AI-assisted diagnostics [23]. Finally, creating specialized, clinically-validated evaluation metrics like FORTE that move beyond linguistic similarity to measure diagnostic fidelity and clinical actionability will be fundamental for translating technical progress into genuine improvements in patient care [7]. As these architectural components continue to mature and integrate more deeply with clinical workflows, MLLMs are poised to become indispensable collaborators in the pursuit of precision neurology and oncology.
Architectures and Applications: How MLLMs are Engineered for Brain MRI Analysis
The application of Multimodal Large Language Models (MLLMs) to 3D medical imaging represents a transformative frontier in medical AI. This guide objectively compares two pioneering architectures—BrainGPT for 3D brain CT report generation and mpLLM for Visual Question Answering (VQA) on multiparametric 3D brain MRI. Framed within broader research on brain MRI sequence classification, this analysis covers their architectural philosophies, experimental performance, and suitability for specific clinical research tasks. Performance data indicates that BrainGPT achieves a Turing test pass rate of 74% and a feature-oriented radiology task evaluation score of 0.71, while mpLLM outperforms baseline models by 5.3% on VQA tasks [7] [27].
BrainGPT: A Holistic Framework for 3D CT Report Generation
BrainGPT is designed to address the critical challenge of generating diagnostically relevant reports from volumetric 3D brain CT scans [7]. Its development involved a comprehensive framework encompassing dataset curation, model fine-tuning, and the creation of a novel evaluation metric.
- Architecture Foundation: Built upon the open-source Otter model, which itself is based on the OpenFlamingo architecture [28]. The model incorporates a CLIP ViT-L/14 visual encoder, a Perceiver Resampler, and an LLaMA-7B language model as its core components [28].
- Clinical Visual Instruction Tuning (CVIT): A key innovation in BrainGPT's training is the use of CVIT, which enhances the model's medical domain knowledge. This includes "keyword instruction," guiding the model to structure reports around specific radiological concepts like Degree, Landmark, Feature, and Impression [7] [28].
- FORTE Evaluation Metric: The authors identified that traditional Natural Language Processing (NLP) metrics were insufficient for evaluating clinical report quality. They developed the Feature-Oriented Radiology Task Evaluation (FORTE), a structured keyword extraction method that assesses the clinical essence of generated reports [7].
mpLLM: A Mixture-of-Experts for Multiparametric MRI VQA
The mpLLM model is engineered to tackle the complexities of Visual Question Answering (VQA) involving multiple, interrelated 3D MRI modalities, a common scenario in clinical practice for diagnosing brain tumors and other intracranial lesions [27].
- Prompt-Conditioned Hierarchical Mixture-of-Experts (MoE): This is the core architectural innovation of mpLLM. It efficiently fuses multiple 3D modalities by routing information through modality-level and token-level projection experts based on the input question. This design dramatically reduces the computational burden compared to simply concatenating vision tokens from all modalities [27] [29].
- Efficient Training without Massive Paired Data: A significant advantage of mpLLM is that it does not require pre-training on large-scale image-report datasets, which are often scarce in medicine. Instead, it can be trained end-to-end on a VQA dataset [27].
- Synthetic VQA Protocol: To overcome the lack of annotated VQA data for 3D brain mpMRI, mpLLM integrates a synthetic data generation protocol that derives medically relevant questions from existing segmentation annotations, which are then clinically validated by medical experts [27].
Table 1: Core Architectural Comparison of BrainGPT and mpLLM
| Feature | BrainGPT | mpLLM |
|---|---|---|
| Primary Task | Automated Report Generation (RRG) [7] | Visual Question Answering (VQA) [27] |
| Imaging Modality | 3D Brain Computed Tomography (CT) [7] | Multiparametric 3D Brain MRI (mpMRI) [27] |
| Core Technical Innovation | Clinical Visual Instruction Tuning (CVIT) [7] | Prompt-Conditioned Hierarchical Mixture-of-Experts (MoE) [27] |
| Data Strategy | Curation of a large-scale dataset (3D-BrainCT: 18,885 text-scan pairs) [7] | Synthetic VQA generation from segmentation masks [27] |
| Key Evaluation Metric | FORTE (Feature-Oriented Radiology Task Evaluation) [7] | Accuracy on expert-validated VQA tasks [27] |
Experimental Protocols and Performance Benchmarking
BrainGPT Experimental Setup and Results
Protocol:
- Dataset: The model was trained and evaluated on the in-house 3D-BrainCT dataset containing 18,885 text-scan pairs [7].
- Fine-Tuning: Four variants of BrainGPT were created by applying different levels of Visual Instruction Tuning (VIT) to the base Otter model: Regular VIT (Plain and In-context Example instructions) and Clinical VIT (Template and Keyword instructions) [7] [28].
- Evaluation: Generated reports were evaluated using both traditional NLP metrics (e.g., BLEU, CIDEr) and the novel FORTE metric. A critical post-processing step of sentence pairing was applied to improve metric alignment. A Turing-like test with 11 physician raters was conducted to assess the clinical indistinguishability of the reports [7].
Performance:
- FORTE Score: The BrainGPT model achieved an average FORTE F1-score of 0.71, with sub-scores for Impression (0.779) and Landmark (0.706) being particularly strong [7].
- Turing Test: In a blinded evaluation, 74% of reports generated by BrainGPT were deemed indistinguishable from those written by human radiologists [7].
- Traditional Metrics: After sentence pairing, the hierarchical improvement from RVIT to CVIT models was best captured by the CIDEr-R metric, which increased from 125.86 (BrainGPT-plain) to 153.3 (BrainGPT-keyword) [7] [28].
mpLLM Experimental Setup and Results
Protocol:
- Dataset: The model was evaluated on the first clinically validated VQA dataset for 3D brain mpMRI, created using its synthetic VQA protocol [27].
- Comparison Baselines: mpLLM was benchmarked against strong medical Vision-Language Model (VLM) baselines [27].
- Ablation Studies: The importance of its core components—modality-level and token-level experts, and prompt-conditioned routing—was validated through ablation experiments [27].
Performance:
- The mpLLM architecture outperformed strong medical VLM baselines by 5.3% on average across multiple mpMRI datasets [27].
Contextual Performance in MRI Sequence Classification
While not their primary function, the capabilities of general MLLMs on tasks like MRI sequence classification provide a useful baseline for understanding the domain's challenges. A recent study evaluated models like ChatGPT-4o, Claude 4 Opus, and Gemini 2.5 Pro on classifying 13 standard brain MRI series [9].
Table 2: Performance Benchmarking Across Tasks and Models
| Model / Task | Key Metric | Reported Score | Context & Notes |
|---|---|---|---|
| BrainGPT (RRG) | FORTE (Avg. F1) | 0.71 [7] | Higher scores indicate better capture of clinical keywords. |
| BrainGPT (Turing Test) | Human Rater Accuracy | 74% [7] | Percentage of reports deemed human-like. |
| mpLLM (VQA) | Accuracy vs. Baselines | +5.3% [27] | Average improvement over other medical VLMs. |
| General MLLMs (Sequence ID) | |||
| ⋄ ChatGPT-4o | Classification Accuracy | 97.7% [9] | On a dataset of 130 brain MRI images. |
| ⋄ Gemini 2.5 Pro | Classification Accuracy | 93.1% [9] | Occasional hallucinations noted [9]. |
| ⋄ Claude 4 Opus | Classification Accuracy | 73.1% [9] | Struggled with SWI and ADC sequences [9]. |
Visualizing Architectures and Workflows
BrainGPT Clinical Fine-Tuning and Evaluation Workflow
mpLLM Mixture-of-Experts Architecture
The Scientist's Toolkit: Essential Research Reagents
For researchers aiming to work in this domain, the following tools and datasets featured in the evaluated models are critical.
Table 3: Key Research Reagents and Resources
| Resource Name | Type | Primary Function in Research | Example Use Case |
|---|---|---|---|
| 3D-BrainCT Dataset [7] | Proprietary Dataset | Provides text-scan pairs for training and evaluating 3D CT report generation models. | Training BrainGPT models [7]. |
| FORTE Metric [7] | Evaluation Metric | Gauges clinical relevance of generated reports by extracting structured keywords (Degree, Landmark, Feature, Impression). | Evaluating diagnostic quality beyond text similarity [7]. |
| Synthetic VQA Protocol [27] | Data Generation Method | Generates medically valid Visual Q&A pairs from segmentation annotations, mitigating data scarcity. | Creating training data for mpLLM without manual VQA annotation [27]. |
| Clinical Visual Instruction Tuning [7] | Training Methodology | Enhances model's clinical reasoning by incorporating structured templates and keyword guidelines during fine-tuning. | Steering BrainGPT to generate clinically sensible reports [7] [28]. |
| Hierarchical MoE Architecture [27] | Model Architecture | Enables parameter-efficient fusion of multiple, interrelated 3D image modalities for joint reasoning. | Allowing mpLLM to process T1w, T2w, and FLAIR MRI sequences effectively [27]. |
| VQA-RAD & ROCOv2 [30] | Public Benchmark Datasets | Standardized datasets for evaluating model performance on medical Visual Question Answering and image captioning. | Benchmarking MLLM performance on clinical tasks [30]. |
The application of Large Language Models (LLMs) in specialized domains like healthcare requires moving beyond general-purpose capabilities to developing nuanced medical expertise. This transition is primarily achieved through three specialized training paradigms: pre-training on domain-specific corpora, instruction tuning to follow task-oriented prompts, and alignment to ensure outputs meet clinical standards. Within the specific context of multimodal LLM (MLLM) performance on brain MRI sequence classification—a critical task for diagnostic assistance and automated report generation—the choice of training strategy significantly impacts model accuracy, reliability, and clinical utility. These paradigms enable the transformation of general foundation models into specialized tools that can interpret complex medical images, classify intricate MRI sequences, and generate clinically coherent radiology reports, thereby addressing one of the most visually and diagnostically challenging tasks in modern radiology.
Comparative Analysis of Training Paradigms
The following table summarizes the core objectives, methodologies, and representative models for each of the three primary training paradigms in the medical domain.
Table 1: Comparison of Core Training Paradigms for Medical LLMs/MLLMs
| Training Paradigm | Primary Objective | Key Methodology | Representative Medical Models |
|---|---|---|---|
| Pre-training | To build foundational medical knowledge and representations from a broad, unlabeled corpus [31] [32]. | Self-supervised learning on large-scale biomedical text (e.g., clinical notes, literature) and images (e.g., X-rays, CTs) [31] [32]. | BioBERT, ClinicalBERT, BiomedGPT (base versions) [33] [32] |
| Instruction Tuning | To adapt models to follow instructions and perform diverse, specific tasks based on natural language prompts [33] [34]. | Supervised fine-tuning on datasets of (instruction, input, output) triplets covering tasks like NER, RE, and QA [33] [35]. | Llama2-MedTuned, Med-Alpaca, BiomedGPT (instruction-tuned) [33] [32] |
| Alignment | To refine model behavior to be helpful, safe, and adhere to clinical standards and constraints [7] [36]. | Human (or AI) feedback on generated outputs (e.g., RLHF), multi-agent review frameworks, and specialized evaluation metrics [7] [36]. | BrainGPT (CVIT), Medical AI Consensus framework [7] [36] |
Experimental Performance in Brain MRI Sequence Classification
Recent benchmarking studies provide quantitative data on the performance of differently trained models on the critical task of brain MRI sequence classification. The table below compares the accuracy of leading multimodal LLMs, highlighting the tangible outcomes of their underlying training approaches.
Table 2: Model Performance on Brain MRI Sequence Classification (n=130 images, 13 series) [9]
| Multimodal LLM | Modality Identification Accuracy | Anatomical Region Recognition Accuracy | Imaging Plane Classification Accuracy | Contrast-Enhancement Status Accuracy | MRI Sequence Classification Accuracy |
|---|---|---|---|---|---|
| ChatGPT-4o | 100% | 100% | 100% | 98.46% | 97.69% |
| Gemini 2.5 Pro | 100% | 100% | 100% | 98.46% | 93.08% |
| Claude 4 Opus | 100% | 100% | 99.23% | 95.38% | 73.08% |
Statistical analysis using Cochran's Q test revealed that the differences in MRI sequence classification accuracy were statistically significant (p < 0.001), underscoring the variable efficacy of different model architectures and training strategies [9]. The most frequent misclassifications involved Fluid-attenuated inversion recovery (FLAIR) sequences, often confused with T1-weighted or diffusion-weighted sequences. Furthermore, Gemini 2.5 Pro exhibited occasional "hallucinations," generating irrelevant clinical details such as "hypoglycemia" and "Susac syndrome," a critical failure mode for clinical deployment [9].
Impact of Specialized Instruction Tuning
Specialized instruction tuning demonstrably enhances model performance on clinical tasks. The Llama2-MedTuned models, instruction-tuned on approximately 200,000 biomedical samples, showed potential to achieve results on par with specialized encoder-only models like BioBERT and BioClinicalBERT for classical biomedical NLP tasks such as Named Entity Recognition (NER) and Relation Extraction (RE) [33]. In radiology report generation, the Clinically Visual Instruction Tuned (CVIT) BrainGPT model, trained on the 3D-BrainCT dataset, achieved an average Feature-Oriented Radiology Task Evaluation (FORTE) F1-score of 0.71. In a Turing-like test, 74% of its generated reports were indistinguishable from human-written ground truth [7].
Detailed Experimental Protocols and Methodologies
Protocol 1: Instruction Tuning for Biomedical NLP
The development of Llama2-MedTuned provides a template for effective instruction tuning [33].
- Dataset Curation: A comprehensive instruction dataset of ~200,000 samples was assembled from diverse public biomedical datasets. This included data for Named Entity Recognition (NCBI-disease, BC5CDR), Relation Extraction (i2b2-2010, GAD), Natural Language Inference (MedNLI), Document Classification (HoC), and Question Answering (ChatDoctor, PMC-Llama).
- Prompt Template: A structured prompt format with three components was adopted: 1) Instruction: One of 5-10 randomly chosen task descriptions; 2) Input: The original text from the source dataset; 3) Output: The expected model response.
- Training Configuration: Models were fine-tuned using the Llama2 (7B and 13B) base models with a batch size of 10 per GPU and DeepSpeed Zero 3 optimization [33].
Protocol 2: Evaluating MLLMs on Brain MRI Classification
A rigorous protocol for evaluating multimodal LLMs on brain MRI classification tasks is detailed in recent comparative studies [9].
- Data Preparation: 130 brain MRI images from adult patients without pathological findings were selected, representing 10 single-slice images for each of 13 standard MRI series (e.g., Axial T1w, T2w, FLAIR, DWI, SWI, ADC, and contrast-enhanced variants). Images were exported in high-quality JPEG format without annotations.
- Prompting and Evaluation: A standardized, zero-shot English prompt was used for all models, asking them to identify the modality, anatomical region, imaging plane, contrast status, and specific MRI sequence. To prevent in-context adaptation, a new chat session was initiated for each query.
- Ground Truth and Analysis: Two radiologists independently reviewed and classified LLM responses as "correct" or "incorrect" in consensus. Accuracy was calculated for each task, and differences in sequence classification performance were analyzed using Cochran's Q test and pairwise McNemar tests with Bonferroni correction [9].
Protocol 3: Clinical Visual Instruction Tuning (CVIT) for Report Generation
The BrainGPT study established a advanced protocol for aligning models with clinical reasoning [7].
- Base Model and Data: The open-source Otter foundation model was used. It was fine-tuned on a curated dataset of 18,885 3D brain CT text-scan pairs.
- Fine-Tuning Conditions: Four distinct instruction-tuning strategies were compared:
- Plain Instruction: Basic role instruction (e.g., "You are a radiology assistant").
- In-Context Example Instruction: Plain instruction supplemented with 3-shot examples.
- Template Instruction: Incorporation of structured clinical QA templates.
- Keyword Instruction: Use of categorical guidelines focused on essential radiology keywords.
- Evaluation Metric - FORTE: A novel Feature-Oriented Radiology Task Evaluation metric was introduced to move beyond traditional NLP scores. FORTE extracts and evaluates four key components from radiology reports:
degree,landmark,feature, andimpression[7].
Workflow and Conceptual Diagrams
Medical MLLM Specialization Pipeline
The following diagram illustrates the end-to-end pipeline for transforming a general-purpose foundation model into a specialized medical MLLM.
Multi-Agent Framework for Evaluation and Alignment
For complex clinical tasks like radiology report generation, a multi-agent framework ensures rigorous evaluation and alignment, as depicted below.
The Scientist's Toolkit: Essential Research Reagents
This section details key datasets, models, and evaluation tools essential for research and development in medical MLLMs for brain MRI analysis.
Table 3: Essential Research Reagents for Medical MLLM Development
| Category | Name | Description and Function |
|---|---|---|
| Datasets | 3D-BrainCT [7] | A curated dataset of 18,885 text-scan pairs of 3D brain CTs. Used for training and evaluating models on volumetric medical image interpretation. |
| Llama2-MedTuned-Instructions [33] | An instruction dataset of ~200,000 samples compiled from various biomedical tasks (NER, RE, NLI, QA). Used for instruction tuning general LLMs for medicine. | |
| Base Models | Llama2 (7B/13B) [33] | A powerful, open-source autoregressive language model. Serves as a common base model for subsequent medical instruction tuning. |
| Otter [7] | An open-source multimodal model. Used as the foundation for clinical visual instruction tuning in the BrainGPT study. | |
| Specialized Models | BiomedGPT-Large/XL [32] | Scaled-up vision-language models (472M/930M parameters). Demonstrate the impact of model scaling on performance across 6 multi-modal biomedical tasks. |
| BrainGPT [7] | A Clinically Visual Instruction Tuned (CVIT) model for 3D CT radiology report generation. Exemplifies the application of advanced instruction tuning. | |
| Evaluation Tools | FORTE [7] | Feature-Oriented Radiology Task Evaluation. A clinical essence metric that evaluates reports on degree, landmark, feature, and impression components. |
| HumanELY [37] | A standardized framework and web app for human evaluation of LLMs in healthcare, addressing metrics like accuracy, harm, and coherence. | |
| Multi-Agent Framework [36] | A benchmark environment with ten specialized agents (e.g., classifier, composer, evaluator) for rigorous radiology report generation and evaluation. |
Multimodal LLM Performance in Brain MRI Sequence Classification
Multimodal Large Language Models (MLLMs) represent a transformative advancement in medical artificial intelligence, combining the reasoning capabilities of large language models with computer vision to interpret complex clinical data. In radiology, these models are increasingly applied to two critical tasks: Visual Question Answering (VQA), which allows interactive querying about image content, and Automated Radiology Report Generation (RRG), which produces preliminary diagnostic reports [3]. The analysis of brain MRI presents particular challenges due to the multiplicity of standard sequences (T1-weighted, T2-weighted, FLAIR, DWI, etc.), each providing complementary clinical information. Accurate sequence classification forms the foundational step toward more complex interpretation tasks, yet this capability varies significantly across current MLLM implementations [9]. This comparison guide examines the current state of MLLM performance specifically for brain MRI sequence classification and related tasks, providing researchers with objective performance data and methodological insights to inform their work.
Performance Comparison Tables
Brain MRI Sequence Classification Accuracy of General-Purpose MLLMs
Table 1: Performance of general-purpose MLLMs on brain MRI sequence classification tasks
| Model | Sequence Classification Accuracy | Contrast-Enhancement Status Accuracy | Imaging Plane Classification Accuracy | Notable Strengths | Common Errors |
|---|---|---|---|---|---|
| ChatGPT-4o | 97.69% (127/130) [9] | 98.46% (128/130) [9] | 100% (130/130) [9] | Excellent sequence recognition, high reliability | Rare misclassification of FLAIR as T1-weighted or DWI |
| Gemini 2.5 Pro | 93.08% (121/130) [9] | 98.46% (128/130) [9] | 100% (130/130) [9] | Strong overall performance | Occasional hallucinations, adding irrelevant clinical details |
| Claude 4 Opus | 73.08% (95/130) [9] | 95.38% (124/130) [9] | 99.23% (129/130) [9] | Competent basic recognition | Struggles with SWI and ADC sequences, lower sequence accuracy |
Specialized Model Performance on 3D Brain MRI Tasks
Table 2: Performance of specialized medical MLLMs on 3D brain MRI tasks
| Model | Primary Task | Key Metric | Performance | Architecture | Clinical Validation |
|---|---|---|---|---|---|
| BrainGPT [7] | 3D CT Report Generation | FORTE F1-Score | 0.71 (average) [7] | Clinical Visual Instruction Tuning (CVIT) | 74% of reports indistinguishable from human in Turing test [7] |
| mpLLM [27] | 3D mpMRI VQA | Average Improvement | Outperforms baselines by 5.3% [27] | Prompt-conditioned hierarchical Mixture-of-Experts | Clinician-validated VQA dataset and model responses |
| MedVersa [38] | Radiology Report Generation | RadCliQ-v1 Score | 1.46 ± 0.03 on IU X-ray findings [38] | Multitask model | Not specified for brain MRI applications |
Experimental Protocols and Methodologies
Brain MRI Sequence Classification Protocol
The comprehensive evaluation of general-purpose MLLMs for brain MRI sequence classification employed a rigorous methodology [9]. Researchers collected 130 brain MRI images from adult patients without pathological findings, representing 13 standard MRI series including axial T1-weighted, T2-weighted, FLAIR (axial, coronal, sagittal), SWI, DWI, ADC, and contrast-enhanced sequences across multiple planes. All images were exported in high-quality JPEG format (minimum resolution 994×1382 pixels) without compression, cropping, or annotations. The study utilized a zero-shot prompting approach with a standardized prompt that asked models to identify: (1) radiological modality, (2) anatomical region, (3) imaging plane, (4) contrast-enhancement status, and (5) specific MRI sequence. To prevent in-context adaptation, researchers initiated a new session for each prompt by clearing chat history. Two radiologists independently reviewed and classified responses as "correct" or "incorrect" through consensus, with hallucinations defined as statements unrelated to the input image or prompt context.
3D Medical Imaging Evaluation Metrics
Specialized models for 3D medical imaging employ tailored evaluation frameworks that address the limitations of traditional natural language processing metrics [7]. The Feature-Oriented Radiology Task Evaluation (FORTE) was specifically developed to capture clinical essence in generated reports by evaluating four essential keyword components: degree, landmark, feature, and impression [7]. This approach recognizes that traditional metrics like BLEU and ROUGE-L correlate poorly with radiologist evaluations [27]. FORTE employs structured keyword extraction that addresses multi-semantic context, recognizes synonyms, and transfers across modalities. Similarly, the S-Score metric evaluates structured reports by measuring both disease prediction accuracy and precision of disease-specific details, demonstrating stronger alignment with human assessments than traditional metrics [39].
Diagram 1: Brain MRI sequence classification experimental workflow. This protocol tests MLLM capability to identify fundamental image characteristics using zero-shot prompting and radiologist consensus validation [9].
Key Architectural Approaches
Specialized Architectures for 3D Medical Imaging
Medical MLLMs employ specialized architectures to address the unique challenges of 3D medical images. BrainGPT utilizes Clinical Visual Instruction Tuning (CVIT) to enhance medical domain knowledge, incorporating four fine-tuning conditions: plain instruction (describing the model's role as a radiology assistant), in-context example instruction (adding 3-shot examples), template instruction (using structured clinical QA templates), and keyword instruction (providing categorical guidelines focused on keywords) [7]. This hierarchical approach progressively enhances clinical sensibility in report generation.
The mpLLM architecture introduces a prompt-conditioned hierarchical Mixture-of-Experts (MoE) specifically designed for multiparametric 3D brain MRI [27]. This approach routes computation across modality-level and token-level projection experts to fuse multiple interrelated 3D modalities efficiently. Unlike modality-specific or modality-agnostic vision encoders, mpLLM's low-level components are lightweight projection functions that train end-to-end with the language model during fine-tuning, dramatically reducing GPU memory usage by processing a single fused vision token representation rather than multiple separate image tokens [27].
Diagram 2: Medical MLLM architecture overview showing standard components (black) and specialized medical adaptations (red). Medical MLLMs build upon general architectures but incorporate domain-specific enhancements like hierarchical MoE and clinical instruction tuning [27] [3].
Table 3: Key research reagents and resources for medical MLLM development
| Resource | Type | Key Features | Research Applications |
|---|---|---|---|
| 3D-BrainCT Dataset [7] | Dataset | 18,885 text-scan pairs of 3D brain CT | Training and evaluation of 3D report generation models |
| MIMIC-STRUC [39] | Structured Dataset | Chest X-ray with disease severity, location, probability | Structured radiology report generation development |
| FORTE [7] | Evaluation Framework | Feature-oriented radiology task evaluation | Clinical essence measurement in generated reports |
| S-Score [39] | Evaluation Metric | Measures disease prediction accuracy and detail precision | Structured report quality assessment |
| ReXGradient-160K [40] | Benchmark Dataset | 273,004 chest X-rays from 160,000 studies | Large-scale RRG and VQA benchmarking |
| Clinical Visual Instruction Tuning [7] | Training Methodology | Enhances medical domain knowledge | Specialized medical MLLM development |
| Hierarchical MoE Architecture [27] | Model Architecture | Efficient fusion of multiple 3D modalities | Multiparametric MRI analysis |
Current MLLMs demonstrate varying capabilities in brain MRI sequence classification and related tasks. General-purpose models like ChatGPT-4o achieve remarkably high accuracy (97.69%) in basic sequence recognition [9], while specialized architectures like BrainGPT and mpLLM address more complex 3D medical imaging challenges through clinical visual instruction tuning and hierarchical mixture-of-experts approaches [7] [27]. The field continues to evolve with improved evaluation metrics like FORTE and S-Score that better capture clinical utility compared to traditional NLP metrics [39] [7]. As these technologies mature, they hold significant promise for enhancing radiologist workflow efficiency and diagnostic consistency, particularly for complex 3D imaging modalities like brain MRI.
The application of Multimodal Large Language Models (MLLMs) to brain MRI analysis represents a frontier in medical artificial intelligence, offering potential breakthroughs in diagnostic accuracy, workflow efficiency, and personalized treatment planning. However, a significant bottleneck hindering progress in this domain is the severe scarcity of high-quality, clinically validated image-text paired datasets for training and evaluation [41]. Unlike natural images, medical data requires specialized expertise for annotation, involves privacy concerns that limit sharing, and encompasses complex 3D volumetric data that standard 2D models cannot effectively process [29] [41].
In response to these challenges, two innovative data solutions have emerged as particularly promising: Synthetic Visual Question Answering (VQA) Generation and Clinical Visual Instruction Tuning (CVIT). These approaches address the data scarcity problem from different angles—synthetic generation creates artificial but medically relevant training data, while CVIT enhances model training through structured, clinically-grounded instruction formats. This guide provides a comprehensive comparison of these methodologies, their experimental protocols, performance outcomes, and practical implementation considerations for researchers and drug development professionals working at the intersection of AI and neuroimaging.
Methodology and Experimental Protocols
Synthetic VQA Generation for 3D Brain MRI
The synthetic VQA approach focuses on algorithmically generating medically relevant question-answer pairs from existing medical image annotations, thereby creating scalable training resources without additional manual clinical labeling.
Experimental Protocol for mpLLM (Multiparametric LLM):
- Data Generation Pipeline: The protocol utilizes existing segmentation annotations from 3D brain MRI datasets to automatically generate medically relevant VQA pairs [29] [42]. This involves creating templates for different question types (e.g., "Is there evidence of [pathology] in the [region]?") and populating them with anatomical and pathological information derived from segmentation masks.
- Clinical Validation: Generated VQA pairs undergo rigorous review by medical experts to ensure clinical relevance and accuracy, creating what researchers describe as "the first clinically validated VQA dataset for 3D brain mpMRI" [29].
- Model Architecture: The mpLLM implements a prompt-conditioned hierarchical mixture-of-experts (MoE) architecture specifically designed to handle multiple interrelated 3D MRI modalities [29] [42]. This includes modality-level experts that process different MRI sequences (T1, T2, FLAIR, etc.) and token-level experts that handle specific regions or features within volumes.
- Training Strategy: The model trains efficiently without image-report pretraining by routing across these specialized experts and leveraging the synthetic VQA data [42]. This approach addresses the limited availability of naturally occurring image-text paired supervision in medical domains.
Clinical Visual Instruction Tuning (CVIT)
CVIT enhances standard visual instruction tuning by incorporating clinical expertise directly into the training process through structured instructions and guidelines.
Experimental Protocol for BrainGPT:
- Instruction Variants: Researchers developed four distinct instruction tuning conditions [41]:
- Plain Instruction: Basic role instruction (model as radiology assistant)
- In-context Example Instruction: 3-shot examples added to plain instruction
- Template Instruction: Structured clinical QA templates added to plain instruction
- Keyword Instruction: Categorical guidelines focused on essential radiology keywords
- Dataset Curation: The approach utilizes the 3D-BrainCT dataset containing 18,885 text-scan pairs, emphasizing lesion diversity including degree, spatial landmarks, and diagnostic impressions [41].
- Evaluation Framework: The protocol introduces Feature-Oriented Radiology Task Evaluation (FORTE), a specialized evaluation scheme that captures clinical essence by focusing on four key components: degree, landmark, feature, and impression [41].
- Model Training: BrainGPT builds upon the Otter foundation model with CVIT enhancements, enabling multi-image captioning capacity with clinical-sensible interpretation of volumetric brain CT scans [41].
Table 1: Core Methodological Differences Between Approaches
| Aspect | Synthetic VQA Generation | Clinical Visual Instruction Tuning (CVIT) |
|---|---|---|
| Primary Innovation | Algorithmic generation of training data from existing annotations | Enhancement of training protocol with clinical expertise |
| Data Requirements | Segmentation masks or anatomical labels | Paired image-text datasets with clinical reports |
| Clinical Validation | Post-generation expert review | Integrated into instruction design |
| Architecture Impact | Requires specialized models (e.g., MoE) for 3D data | Compatible with various base architectures |
| Evaluation Focus | Accuracy on medically relevant VQA tasks | Clinical utility and information fidelity (via FORTE) |
Performance Comparison and Benchmark Results
Quantitative Performance Metrics
Both synthetic VQA and CVIT approaches demonstrate significant performance improvements over baseline models, though they excel in different aspects of medical MLLM capabilities.
Table 2: Performance Comparison Across Methodologies
| Model/Methodology | Dataset/Task | Performance Metrics | Baseline Comparison |
|---|---|---|---|
| mpLLM (Synthetic VQA) | Multiple mpMRI datasets | +5.3% average improvement on medical VQA | Outperforms strong medical VLM baselines by significant margin [29] |
| BrainGPT (CVIT) | 3D-BrainCT Report Generation | FORTE F1-score: 0.71 (Degree: 0.661, Landmark: 0.706, Feature: 0.693, Impression: 0.779) | Superior to baseline Otter model (low BLEU-4 and CIDEr-R scores) [41] |
| BrainGPT (CVIT) | Turing Test Evaluation | 74% of reports indistinguishable from human-written ground truth | Demonstrates clinical quality matching human performance [41] |
| RadFM | RadBench Comprehensive Evaluation | Outperforms GPT-4V and other accessible multimodal foundation models | Strong performance across diagnosis, VQA, and report generation tasks [43] |
Task-Specific Performance Analysis
The comparative advantage of each approach becomes evident when examining performance across different clinical tasks:
Visual Question Answering Performance: Models leveraging synthetic VQA generation, particularly mpLLM, demonstrate strong performance on structured question-answering tasks involving multiparametric MRI data [29]. The hierarchical mixture-of-experts architecture enables effective routing across different MRI modalities, making it particularly suitable for complex diagnostic queries requiring integration of multiple image types.
Report Generation Quality: CVIT-enhanced models like BrainGPT excel in radiology report generation, with 74% of generated reports being indistinguishable from human-written ground truth in Turing-like evaluations [41]. The template and keyword instruction variants show particularly strong performance in generating clinically structured reports with appropriate terminology.
Clinical Utility Assessment: Traditional NLP metrics like BLEU and ROUGE show poor correlation with clinical utility in brain MRI report evaluation [41]. The FORTE evaluation framework demonstrates that CVIT-enhanced models achieve substantially better performance on clinically essential elements including lesion degree (0.661), landmark localization (0.706), feature description (0.693), and diagnostic impression (0.779) [41].
Implementation Considerations and Research Reagents
Essential Research Reagent Solutions
Successful implementation of synthetic VQA generation or CVIT requires specific technical components and research reagents.
Table 3: Essential Research Reagents for Implementation
| Research Reagent | Function | Implementation Examples |
|---|---|---|
| Clinically Validated Datasets | Provide ground truth for training and evaluation | 3D-BrainCT (18,885 text-scan pairs) [41], MedMD (16M 2D/3D radiology scans) [43] |
| Specialized Model Architectures | Handle 3D medical data and clinical reasoning | Prompt-conditioned hierarchical MoE [29], Clinical Visual Instruction Tuning framework [41] |
| Evaluation Frameworks | Assess clinical relevance beyond traditional metrics | FORTE (Feature-Oriented Radiology Task Evaluation) [41], RadBench comprehensive benchmark [43] |
| Data Augmentation Tools | Generate synthetic training data | Synthetic VQA protocol from segmentation annotations [29], Retinex-based image enhancement [44] |
| Clinical Validation Protocols | Ensure medical accuracy and safety | Expert review cycles [29], Turing-like linguistic evaluation [41] |
Workflow Integration Diagrams
The comparative analysis of synthetic VQA generation and clinical visual instruction tuning reveals complementary strengths that can inform research directions in brain MRI MLLM development.
Synthetic VQA generation demonstrates particular value for scenarios with limited textual annotations but available segmentation masks or anatomical labels. Its scalability and ability to generate large training datasets make it suitable for establishing baseline capabilities across diverse MRI modalities and anatomical regions. The approach shows strong performance on structured VQA tasks and enables efficient training without extensive image-report pretraining [29] [42].
Clinical Visual Instruction Tuning excels in applications requiring high-quality report generation and clinical decision support. The structured incorporation of clinical expertise through templates and keyword guidance produces models whose outputs are largely indistinguishable from human-generated reports [41]. CVIT-enhanced models demonstrate superior performance on clinically nuanced tasks requiring accurate lesion characterization, spatial localization, and diagnostic impression formulation.
For research teams and drug development professionals, the choice between these approaches should be guided by specific application requirements, available data resources, and clinical use case priorities. Teams with strong clinical collaboration access may leverage CVIT for superior report generation, while those with extensive image libraries but limited text annotations may benefit from synthetic VQA approaches. Ultimately, the most promising direction may involve hybrid methodologies that combine the scalability of synthetic data generation with the clinical grounding of structured instruction tuning.
Future research should focus on standardizing evaluation metrics across studies, improving generalization to real-world clinical data, and developing more sophisticated synthetic data generation techniques that capture the full complexity of clinical reasoning in neuroimaging.
Navigating Challenges: Strategies to Optimize MLLM Performance and Mitigate Pitfalls
In the pursuit of advanced artificial intelligence systems capable of interpreting complex multimodal data, hallucination remains a fundamental barrier to reliability and trust. Hallucinations in Multimodal Large Language Models (MLLMs) refer to generated outputs that contradict the visual or textual input data, creating significant challenges for real-world deployment [45] [46]. Within medical imaging applications, particularly brain MRI sequence classification, these hallucinations manifest as incorrect classifications, fabricated image interpretations, or misidentified anatomical structures that could directly impact diagnostic accuracy and patient care [47] [48]. The consequences are particularly severe in clinical environments, where such errors could lead to misdiagnosis, inappropriate treatment planning, or overlooked pathological findings [18]. This comprehensive analysis examines the root causes, evaluates the consequences, and compares state-of-the-art correction mechanisms for hallucinations in multimodal AI systems, with special emphasis on their implications for brain MRI research and clinical applications.
Understanding the Causes: Multimodal Disconnects and Architectural Limitations
Hallucinations in MLLMs stem from complex interactions between data quality, model architecture, and training methodologies. Research has identified several primary causation categories that contribute to unreliable outputs in multimodal systems.
Data-Driven and Architectural Origins
Data Quality Issues: Models trained on low-quality, imbalanced, or improperly labeled datasets exhibit heightened hallucination rates due to insufficient contextual learning [46]. In medical imaging contexts, this includes insufficiently diverse pathological representations or inconsistent labeling protocols across institutions [47].
Vision Encoder Limitations: Visual processing components may introduce errors through inadequate feature extraction or poor handling of visual nuances, particularly in noisy or ambiguous contexts [46]. For brain MRI analysis, this could manifest as failure to capture subtle pathological signatures or artifacts misinterpreted as anatomical features [48].
Cross-Modal Alignment Failures: Misalignment between different data modalities—such as temporal mismatches between image sequences and textual descriptions—creates inconsistent internal representations that generate conflicting outputs [46].
Language Prior Over-Reliance: The strong linguistic priors in LLM components often suppress visual evidence in favor of text-based patterns, leading to factual inconsistencies [49] [50]. This is particularly problematic in specialized domains like neuroimaging, where technical terminology may diverge from common usage.
Medical Imaging-Specific Challenges
Brain MRI classification introduces domain-specific hallucination triggers. Domain shift between training and deployment environments—such as when models trained on adult MRI data are applied to pediatric cases—represents a significant challenge [48]. One study demonstrated that ResNet-18 experienced performance degradation when applied to pediatric neuroimaging data after being trained exclusively on adult scans, though hybrid architectures like MedViT showed improved robustness with accuracy improvements from 0.893 to 0.905 after expert domain knowledge adjustments [48]. Additionally, adversarial attacks present serious concerns; research shows that deliberately fabricated details in clinical prompts can trigger hallucination rates between 50-82% across various LLMs, highlighting the vulnerability of these systems in clinical decision support contexts [18].
Correction Mechanisms: Comparative Analysis of Mitigation Strategies
Recent research has produced diverse approaches to mitigating hallucinations in MLLMs, ranging from architectural modifications to inference-time interventions. The table below provides a systematic comparison of prominent mitigation methods evaluated through standardized benchmarks.
Table 1: Comparative Analysis of MLLM Hallucination Mitigation Methods
| Method | Core Approach | Inference Overhead | Reported Efficacy | Key Advantages |
|---|---|---|---|---|
| D-LEAF [49] | Dynamic layer-wise attention diagnostics and correction | ~8% throughput drop | 53% reduction in hallucination rates; ~4% improvement in VQA accuracy/F1-score | Precise head-level intervention preserves correct attention patterns |
| DeCo [50] | Dynamic correction decoding integrating knowledge from preceding layers | Moderate | Significant reduction in hallucination rates across multiple benchmarks | Model-agnostic; compatible with various decoding strategies |
| Bottom-Up Holistic Reasoning [51] | Verifies and integrates perception-level information with cognition-level knowledge | Not specified | Significant improvements across multiple hallucination benchmarks | Addresses both perception and cognition-level hallucinations |
| Training-based Mitigation [46] | Visual instruction tuning, RLHF, and curated dataset training | None after deployment | Variable; dependent on training data quality and volume | Permanent model improvement without runtime costs |
| Prompt-based Mitigation [18] | Specialized prompting to use only clinically validated information | None | Reduces hallucination rate from 66% to 44% on average | Immediately deployable without model retraining |
Specialized Mechanisms for Medical Imaging
In brain MRI classification tasks, specialized approaches have emerged to address domain-specific challenges. Hybrid architectures combining CNNs with transformer components have demonstrated remarkable effectiveness against domain shift problems. The MedViT model, which integrates convolutional layers with self-attention mechanisms, achieved 0.905 accuracy (95% CI 0.893-0.916) on pediatric MRI data after expert domain knowledge adjustments, significantly outperforming ResNet-18 alone [48]. This architecture leverages local feature extraction capabilities of CNNs with the global contextual understanding of transformers, creating more robust representations less susceptible to hallucinatory outputs when confronted with unfamiliar imaging protocols or patient populations.
Additionally, expert domain knowledge integration has proven valuable for medical imaging applications. This approach involves adjusting model decision processes to align with clinical expertise, such as ignoring implausible classifications based on anatomical constraints or known imaging characteristics [48]. For instance, excluding neurologically impossible sequence predictions based on scan orientation or anatomical coverage helps prevent clearly hallucinated outputs that violate basic neuroanatomical principles.
Experimental Protocols: Methodologies for Hallucination Assessment
Rigorous evaluation protocols are essential for quantifying hallucination rates and mitigation effectiveness. Standardized benchmarks and assessment methodologies have been developed across the research community.
Benchmarking and Evaluation Metrics
Comprehensive hallucination assessment typically employs multiple complementary benchmarks to evaluate different aspects of model performance. For general MLLM evaluation, benchmarks such as MMHal and CHAIR assess object-level hallucination rates in generated descriptions [49]. In medical contexts, specialized evaluations include adversarial vignette testing, where models are presented with clinical cases containing deliberately fabricated elements to measure their propensity for elaborating on false information [18].
Standard evaluation metrics include:
- Hallucination Rate: Percentage of generated outputs containing elements unsupported by input data
- Accuracy and F1-Score: Standard classification metrics, particularly for visual question answering tasks
- Temporal Consistency: For video-language models, measures coherence across sequential frames
- Domain Shift Robustness: Performance retention when applying models to data distributions different from training sets
Protocol Implementation Details
The D-LEAF methodology employs Layer Image Attention Entropy (LIAE) to flag anomalous layers and Image Attention Focus (IAF) to identify specific attention heads requiring correction [49]. This approach enables precise, localized interventions rather than blanket suppression of attention mechanisms. Experiments typically evaluate performance across three representative MLLM architectures on three standard multimodal hallucination benchmarks with comparison against multiple state-of-the-art correction methods [49].
For medical imaging applications, protocols often involve retrospective multicenter studies with expert-validated labels. One comprehensive study utilized 8544 examinations and 63,327 sequences from 249 hospitals to develop and evaluate classification models, with rigorous quality assessment excluding sequences with artifacts or unclear labels [48]. This large-scale validation is essential for establishing clinical reliability.
Research Reagent Solutions: Essential Tools for Hallucination Research
Table 2: Essential Research Reagents for Hallucination Investigation and Mitigation
| Reagent / Tool | Function | Example Implementations |
|---|---|---|
| Benchmark Datasets | Standardized evaluation of hallucination rates | MMHal, CHAIR, adversarial clinical vignettes [49] [18] |
| Attention Diagnostic Tools | Identify problematic layers and attention heads | LIAE (Layer Image Attention Entropy), IAF (Image Attention Focus) [49] |
| Hybrid Architectures | Enhanced robustness to domain shift | MedViT (CNN-Transformer hybrid), ResNet-18 with transformer layers [48] |
| Contrastive Decoding Methods | Suppress hallucinated content by comparing distributions | VCD (contrasts original/distorted visual inputs), MoLE (Mixture of Experts) [49] |
| Expert Validation Frameworks | Clinical verification of model outputs | Physician-validated simulated vignettes, multi-reader consensus protocols [18] |
Visualizing the Diagnostic and Correction Workflow
The D-LEAF framework implements a sophisticated diagnostic and correction pipeline that dynamically localizes and addresses hallucination sources during inference. The following diagram illustrates this comprehensive workflow:
Diagram 1: D-LEAF Attention Diagnostic and Correction Workflow
The systematic mitigation of hallucinations in multimodal LLMs represents a critical advancement pathway for reliable medical imaging AI. Current research demonstrates that targeted, mechanistic approaches like D-LEAF and specialized architectures like MedViT significantly outperform blanket suppression methods, offering substantial improvements in output fidelity while maintaining computational efficiency [49] [48]. For brain MRI sequence classification and broader clinical applications, the integration of domain-specific knowledge with dynamic inference-time corrections creates promising avenues for developing robust systems resistant to domain shift and adversarial attacks [48] [18].
The progression toward clinically dependable AI requires continued emphasis on rigorous benchmarking, transparent model interpretability, and collaborative validation between AI researchers and clinical experts. Only through such comprehensive approaches can multimodal systems achieve the reliability standards necessary for meaningful integration into clinical workflows and research applications.
This guide objectively compares modern techniques for overcoming data limitations in paired image-text datasets, framed within multimodal Large Language Model (MLLM) performance for brain MRI sequence classification. We synthesize experimental data from recent research to provide a clear comparison of dataset distillation, data augmentation, and specialized architectural approaches. The analysis reveals that dataset distillation can nearly double retrieval performance with just 100 training pairs, while advanced augmentation significantly enhances model robustness in clinical settings. Supporting data, detailed experimental protocols, and essential research resources are provided to facilitate implementation for researchers and drug development professionals working with constrained multimodal data.
Multimodal Large Language Models (MLLMs) represent a transformative advancement in medical artificial intelligence, particularly for visually intensive disciplines like radiology where they show promise in enhancing diagnostic accuracy and clinical decision-making [9]. However, their effective application to specialized domains such as brain MRI sequence classification faces a fundamental constraint: the limited availability of high-quality, expertly annotated paired image-text datasets. In radiology, an MLLM that cannot reliably recognize fundamental image characteristics like sequence type cannot be expected to reliably analyze clinically complex scenarios [9].
The challenge is particularly pronounced for 3D medical images like brain CTs and MRIs, where traditional multimodal approaches have focused primarily on 2D images, leaving 3D spatial modality interpretation largely unexplored [7]. This guide systematically compares the most effective techniques for overcoming these data limitations, with direct application to brain MRI classification research.
Comparative Analysis of Techniques
Dataset Distillation for Vision-Language Pairs
Dataset distillation addresses data scarcity by creating compact, information-dense versions of large datasets that preserve essential training information. For multimodal medical data, this technique is particularly valuable as it can reduce the number of required training pairs by orders of magnitude while maintaining diagnostic accuracy.
Recent research has developed the first vision-language dataset distillation method, building on trajectory matching to jointly distill image-text pairs in a contrastive formulation [52]. This approach specifically addresses the challenge that vision-language datasets lack discrete classes, instead containing intricate relationships between visual and textual elements.
Experimental Protocol: The evaluation used standard Flickr30K and COCO retrieval benchmarks. The core method involves:
- Modeling relationships between images and corresponding text descriptions
- Updating the distilled dataset to reflect model learning progress during training
- Using Low-Rank Adaptation (LoRA) matching for efficient trajectory matching
- Comparing against coreset selection methods (Herding, K-center, Forgetting)
Table 1: Performance Comparison of Dataset Distillation Methods on Retrieval Tasks
| Method | Training Pairs | Image-to-Text Retrieval Accuracy (Recall@1) | Performance vs. Full Dataset |
|---|---|---|---|
| Coreset Selection (Best) | 1000 | 5.6% | ~10% |
| Vision-Language Distillation | 100 | 9.9% | ~18% |
| Full Dataset Training | ~30,000 | 54.2% | 100% |
The results demonstrate that the distillation approach almost doubles the retrieval performance compared to the best coreset selection method while using an order of magnitude fewer training pairs [52]. This has significant implications for brain MRI research, where collecting large datasets is often impractical.
Data Augmentation Techniques for Multimodal Data
Data augmentation creates modified versions of existing data to improve model training without collecting new samples. For medical multimodal applications, the key is applying synchronized transformations that maintain alignment between images and their corresponding text [53].
Experimental Protocol for Medical Imaging:
- Image Transformations: Apply orientation adjustments (flipping, rotation), resizing (scaling, cropping), perspective adjustments, and color/lighting modifications
- Text Synchronization: Update corresponding captions or regenerate descriptions using models like BLIP or Flamingo
- Validation: Ensure augmented pairs maintain clinical accuracy and anatomical consistency
Table 2: Efficacy of Data Augmentation Techniques in Medical Imaging
| Technique | Application Context | Performance Improvement | Limitations |
|---|---|---|---|
| Geometric Transformations (flip, rotate) | General object recognition | ~5% AUC increase [53] | Risk of anatomical implausibility |
| CutMix/MixUp | Object detection, rare classes | 23% accuracy gain in product recognition [53] | May create clinically unrealistic composites |
| GAN-Based Synthesis | Medical imaging, rare classes | Effective for data diversification [53] | Computationally intensive, requires validation |
| Color/Lighting Adjustments | Generalization across devices | Improved adaptability [54] | Limited impact on structural understanding |
In one real-world application, random cropping with different aspect ratios provided a 23% accuracy increase in recognizing technical product photos compared to using just flips and rotations [53]. For brain MRI classification, similar principles apply but require careful consideration of anatomical plausibility.
Specialized Multimodal Architecture Designs
Model architecture choices significantly impact data efficiency. The Clinical Visual Instruction Tuning (CVIT) approach enhances medical domain knowledge by incorporating structured clinical templates and categorical keyword guidelines during fine-tuning [7].
Experimental Protocol for BrainGPT:
- Curate specialized dataset (3D-BrainCT: 18,885 text-scan pairs)
- Implement four fine-tuning conditions with increasing clinical guidance
- Evaluate using Feature-Oriented Radiology Task Evaluation (FORTE)
- Conduct Turing-like tests with physician raters
Results showed that 74% of BrainGPT-generated reports were indistinguishable from human-written ground truth, with FORTE F1-scores reaching 0.71 for specialized models [7]. This demonstrates how domain-specific architectural adjustments can compensate for data limitations.
Experimental Protocols and Methodologies
Dataset Distillation Implementation
The vision-language dataset distillation method employs these key steps [52]:
- Relationship Modeling: Capture semantic correlations between image patches and text tokens using contrastive learning objectives
- Trajectory Matching: Align learning trajectories between models trained on full and distilled datasets
- Synthetic Example Generation: Create informative synthetic pairs rather than selecting existing samples
- LoRA Integration: Apply Low-Rank Adaptation for efficient parameter updates during matching
The process generates a small set of synthetic image-text pairs that encapsulate the essential information from the original large dataset, enabling efficient model training.
Multimodal Data Augmentation Workflows
For brain MRI applications, augmentation requires specialized protocols [53]:
- Modality-Specific Transformations: Apply clinically plausible modifications to MRI sequences
- Text Alignment: Update corresponding descriptions using rule-based systems or generative models
- Clinical Validation: Verify that augmented samples maintain diagnostic integrity through expert review
- Synchronization Checks: Ensure temporal and spatial alignment in multimodal pairs
Critical considerations include avoiding anatomically impossible transformations and preserving pathological features during augmentation.
Diagram 1: Multimodal Data Augmentation Workflow - This workflow illustrates the parallel processing of image and text data with synchronization and clinical validation checkpoints.
Performance Comparison in Brain MRI Classification
Recent comparative analysis of multimodal LLMs in brain MRI sequence classification provides concrete performance data [9]:
Table 3: Multimodal LLM Performance on Brain MRI Classification Tasks
| Model | Modality Identification Accuracy | Anatomical Region Accuracy | MRI Sequence Classification Accuracy |
|---|---|---|---|
| ChatGPT-4o | 100% | 100% | 97.7% |
| Gemini 2.5 Pro | 100% | 100% | 93.1% |
| Claude 4 Opus | 100% | 100% | 73.1% |
The study used 130 brain MRI images representing 13 standard MRI series, with evaluations conducted in zero-shot settings [9]. Notably, misclassifications most frequently involved fluid-attenuated inversion recovery (FLAIR) sequences, often confused with T1-weighted or diffusion-weighted sequences. These performance differences highlight how architectural choices impact data efficiency in specialized medical tasks.
Table 4: Research Reagent Solutions for Multimodal Medical AI
| Resource Category | Specific Tools/Datasets | Function and Application |
|---|---|---|
| Multimodal Datasets | 3D-BrainCT (18,885 pairs) [7], Flickr30K [52], MS-COCO [55] | Benchmarking and training vision-language models |
| Data Augmentation Libraries | Albumentations, torchvision, nlpaug [53] | Applying modality-specific transformations |
| Evaluation Frameworks | FORTE (Feature-Oriented Radiology Task Evaluation) [7] | Gauging clinical essence in generated reports |
| Model Architectures | BrainGPT [7], CLIP [55], BLIP [53] | Specialized models for multimodal medical data |
| Distillation Tools | Vision-Language Distillation Framework [52] | Creating compact, informative training sets |
Diagram 2: Technical Approaches to Data Limitations - This diagram shows the three primary technical strategies and their integration points for addressing data constraints in multimodal learning.
The comparative analysis reveals that dataset distillation currently offers the most promising approach for data-efficient multimodal learning in brain MRI classification, nearly doubling retrieval performance with just 100 training pairs compared to conventional selection methods [52]. However, optimal results likely require combining these techniques—using distillation to create compact datasets, augmentation to increase variability, and specialized architectures like BrainGPT to incorporate domain knowledge [7].
Future research should address computational intensity in distillation methods and develop more sophisticated evaluation metrics like FORTE that better capture clinical relevance beyond traditional n-gram matching [7]. As multimodal LLMs continue to evolve, these data-efficient techniques will play a crucial role in making advanced AI accessible for specialized medical applications where large datasets remain impractical to collect.
Multimodal Large Language Models (MLLMs) represent a transformative advancement in artificial intelligence, with particular promise in visually intensive medical disciplines such as neuroradiology. These models can process and synthesize information across different data types, including medical images and clinical text. However, their diagnostic performance is not merely a function of their architectural sophistication or training data volume. Rather, prompt engineering—the strategic construction and combination of input elements—emerges as a critical determinant of diagnostic accuracy. This review synthesizes recent evidence from brain MRI research to analyze how specific multimodal prompt elements function as powerful levers, drastically altering the clinical utility of MLLMs in diagnostic classification and differential diagnosis.
Experimental Insights into Prompt Element Efficacy
Quantitative Impact of Individual Prompt Components
Recent research systematically investigates how different combinations of input elements affect the diagnostic performance of models like GPT-4V in challenging brain MRI cases. These studies typically deconstruct the prompt into core components: the unannotated Image (I), expert Annotations (A) such as arrows highlighting pathology, the patient's Medical History (H), and a textual Image Description (D) detailing radiological findings [56] [57].
Table 1: Diagnostic Accuracy of GPT-4V with Different Input Combinations [56]
| Input Combination | Binary Diagnostic Accuracy (%) |
|---|---|
| Image (I) alone | 2.2% |
| Image + Annotations (I+A) | 1.1% |
| Image + Annotations + Medical History (I+A+H) | 36.1% |
| Image + Annotations + Medical History + Image Description (I+A+H+D) | 69.0% |
The data reveals a striking hierarchy of value among input elements. Providing the model with only the image, or the image with basic annotations, yields remarkably low diagnostic accuracy (~1-2%), barely better than random guessing for complex diagnoses [56]. The introduction of structured textual elements dramatically shifts this performance. Regression analyses from these studies quantify the individual contributions, identifying the textual Image Description (D) as the strongest positive predictor of accuracy (Odds Ratio: 68.03, p < 0.001), followed by the patient's Medical History (H) (Odds Ratio: 4.18, p < 0.001) [56]. The visual elements (I and A), while necessary, proved insufficient alone, acting as weak carriers of diagnostic signal without semantic clarification from text.
Comparative Performance of Leading MLLMs on Foundational Tasks
Beyond differential diagnosis, the foundational ability of MLLMs to recognize basic MRI characteristics is a prerequisite for reliable clinical application. A 2025 comparative analysis evaluated three advanced models—ChatGPT-4o, Claude 4 Opus, and Gemini 2.5 Pro—on a brain MRI sequence classification task, a critical first step in image interpretation [9].
Table 2: MLLM Accuracy in Brain MRI Sequence Classification (n=130 images) [9]
| Model | Modality Identification | Anatomical Region | Imaging Plane | Contrast Status | MRI Sequence Classification |
|---|---|---|---|---|---|
| ChatGPT-4o | 100% | 100% | 100% | 98.46% | 97.69% |
| Gemini 2.5 Pro | 100% | 100% | 100% | 98.46% | 93.08% |
| Claude 4 Opus | 100% | 100% | 99.23% | 95.38% | 73.08% |
All models achieved perfect or near-perfect scores on basic recognition tasks (modality, anatomy). However, performance diverged significantly on the more specialized task of identifying the specific MRI sequence, with accuracy ranging from 97.7% for ChatGPT-4o down to 73.1% for Claude 4 Opus [9]. This indicates that while MLLMs are robust in general image understanding, their domain-specific knowledge bases and architectures differ considerably. The study also noted that hallucinations—such as Gemini 2.5 Pro inventing irrelevant clinical details like "hypoglycemia"—remain a critical challenge for clinical deployment [9].
Detailed Experimental Protocols in Multimodal Brain MRI Research
Protocol 1: Evaluating Prompt Elements in Differential Diagnosis
A seminal study by Schramm et al. (2025) established a rigorous protocol to isolate the effect of prompt engineering on diagnostic performance [56] [57].
- Case Selection: Sixty brain MRI cases with challenging yet definitively verified diagnoses were selected. This ensured the task required nuanced reasoning beyond simple pattern recognition.
- Prompt Design: Seven distinct prompt groups were systematically constructed from combinations of four core elements: the unannotated Image (I), expert Annotations (A) highlighting key findings, the patient's Medical History (H), and a textual Image Description (D) of radiological findings.
- Query and Evaluation: For each case and prompt group, three identical queries were performed using an LLM-based search engine (Perplexity AI, powered by GPT-4V). The generated differential diagnoses were evaluated against the ground truth using both a binary (correct/incorrect) and a numeric scoring system (ranking the correct diagnosis 1st, 2nd, or 3rd). Statistical analyses included chi-square and Kruskal-Wallis tests, corrected for false-discovery rate [56].
Protocol 2: Benchmarking MLLMs on Sequence Classification
A 2025 study by PMC provided a protocol for evaluating the core visual recognition capabilities of MLLMs, a foundational layer for diagnosis [9].
- Dataset Curation: 130 brain MRI images from adult patients without pathological findings were used, representing 10 single-slice images for each of 13 standard MRI series (e.g., T1w, T2w, FLAIR, DWI, SWI).
- Zero-Shot Prompting: Each image was uploaded individually to the models (ChatGPT-4o, Claude 4 Opus, Gemini 2.5 Pro) with a standardized, zero-shot prompt. The prompt explicitly stated the context was for medical research and requested identification of the modality, anatomical region, imaging plane, contrast status, and specific MRI sequence.
- Analysis: Responses were independently reviewed by two radiologists in consensus and classified as correct or incorrect. Accuracy was calculated for each task, and differences in sequence classification performance were analyzed using Cochran's Q test and pairwise McNemar tests with Bonferroni correction [9].
Protocol 3: Human-AI Collaboration Workflow
A 2025 usability study explored a more realistic clinical scenario: radiology residents using an LLM as an adjunct tool for brain MRI differential diagnosis [58].
- Study Design: Six radiology residents read forty challenging brain MRI cases in a randomized, crossover design. For twenty cases, they used a conventional internet search, and for the other twenty, they used an LLM-assisted search engine (Perplexity AI with GPT-4).
- Measurement: Participants provided their top three differential diagnoses for each case. The primary outcome was diagnostic accuracy, with secondary metrics including interpretation time and confidence level.
- Result: The LLM-assisted workflow yielded significantly superior accuracy (61.4%) compared to the conventional workflow (46.5%). The study also provided crucial insights into human-AI interaction challenges, finding that correct LLM suggestions were adopted by readers in 82.1% of cases, but also identifying LLM hallucinations (11.5% of cases) and inaccurate user descriptions of image findings as key pitfalls [58].
Visualizing the Multimodal Prompt Engineering Workflow
The following diagram illustrates the logical pathway and profound impact of integrating different input elements on the diagnostic output of an MLLM, as revealed by the experimental data.
Table 3: Key Reagents and Resources for Brain MRI MLLM Research
| Resource | Type | Function in Research |
|---|---|---|
| Challenging Brain MRI Datasets [56] [57] | Curated Image Sets | Provides verified, complex cases with definitive diagnoses to test diagnostic reasoning beyond simple recognition. |
| Structured Prompt Groups [56] | Experimental Protocol | Enables systematic isolation and measurement of the contribution of individual input elements (I, A, H, D) to performance. |
| OmniBrainBench [6] | Comprehensive Benchmark | Evaluates MLLMs across the full clinical continuum (15 modalities, 15 tasks), highlighting gaps in complex reasoning. |
| FORTE (Feature-Oriented Radiology Task Evaluation) [7] | Evaluation Metric | Gauges the clinical essence of generated reports by extracting and scoring keywords related to degree, landmark, feature, and impression. |
| MMRQA Framework [8] | Quality Assessment Tool | Integrates quantitative MRI signal metrics (SNR, CNR) with MLLM semantic reasoning for interpretable image quality assessment. |
| Human-AI Collaboration Usability Protocol [58] | Study Design | Measures real-world diagnostic impact and identifies interaction challenges like automation bias and prompt inaccuracy. |
The empirical evidence is clear: in the application of Multimodal LLMs to brain MRI analysis, prompt engineering is not a minor optimization but a fundamental determinant of diagnostic performance. The drastic leap from near-useless (1-2%) to clinically suggestive (69%) accuracy hinges on the strategic inclusion of structured textual elements—specifically, semantic image descriptions and medical history [56]. While leading models like ChatGPT-4o demonstrate impressive foundational capabilities in sequence classification [9], their effective integration into the diagnostic workflow, particularly in a human-AI collaborative model [58], requires meticulously engineered prompts that bridge the gap between visual data and clinical context. Future research must continue to refine these engineering principles, develop robust evaluation benchmarks like OmniBrainBench [6], and prioritize mitigation of hallucinations to responsibly realize the full potential of MLLMs in clinical neuroscience.
The integration of Multimodal Large Language Models (MLLMs) into clinical practice, particularly for specialized tasks like brain MRI sequence classification, presents a dual challenge: achieving high diagnostic accuracy while ensuring robust data privacy and computational efficiency. Brain MRI interpretation requires precise recognition of imaging sequences—a foundational task where models must correctly identify sequences like T1-weighted, T2-weighted, FLAIR, and DWI to provide clinically reliable interpretations [9]. Recent evaluations of general-purpose MLLMs reveal significant performance variations in this specific domain, with accuracy ranging from 73.1% to 97.7% for sequence classification, highlighting a critical performance gap that must be addressed before reliable clinical deployment [9].
Two technological paradigms have emerged to bridge this gap: Retrieval-Augmented Generation (RAG) and Local Fine-Tuning. RAG enhances model accuracy by dynamically integrating external, authoritative knowledge sources without modifying the underlying model, while local fine-tuning adapts model weights to specific clinical domains using institutional data. When deployed on-premises or through hybrid architectures, these approaches simultaneously address computational and privacy concerns inherent in healthcare environments governed by regulations like HIPAA. This analysis examines the comparative performance, implementation methodologies, and optimization strategies for these approaches within the specific context of brain MRI sequence classification research.
Experimental Foundation: Benchmarking MLLM Performance on Brain MRI Classification
Quantitative Performance Assessment of State-of-the-Art MLLMs
A rigorous 2025 benchmark study evaluated three advanced MLLMs—ChatGPT-4o, Claude 4 Opus, and Gemini 2.5 Pro—on their ability to classify brain MRI sequences using 130 expertly annotated brain MRI images representing 13 standard sequences [9]. The evaluation assessed five critical classification tasks, with MRI sequence identification serving as the primary outcome measure. The results demonstrate substantial performance variation across models, with ChatGPT-4o achieving superior accuracy in the most clinically demanding task [9].
Table 1: Performance Comparison of Multimodal LLMs on Brain MRI Classification Tasks (n=130 images)
| Model | Modality Identification | Anatomical Region Recognition | Imaging Plane Classification | Contrast-Enhancement Status | MRI Sequence Classification |
|---|---|---|---|---|---|
| ChatGPT-4o | 130/130 (100%) | 130/130 (100%) | 130/130 (100%) | 128/130 (98.46%) | 127/130 (97.69%) |
| Gemini 2.5 Pro | 130/130 (100%) | 130/130 (100%) | 130/130 (100%) | 128/130 (98.46%) | 121/130 (93.08%) |
| Claude 4 Opus | 130/130 (100%) | 130/130 (100%) | 129/130 (99.23%) | 124/130 (95.38%) | 95/130 (73.08%) |
Statistical analysis using Cochran's Q test revealed significant differences in MRI sequence classification performance (p < 0.001), establishing this task as a key differentiator for clinical readiness [9]. The most frequent misclassifications involved Fluid-Attenuated Inversion Recovery (FLAIR) sequences, often confused with T1-weighted or diffusion-weighted sequences, indicating specific areas requiring model improvement [9].
Methodological Framework for Brain MRI Sequence Classification
The benchmark study employed a standardized experimental protocol to ensure rigorous evaluation [9]:
Image Selection and Preparation: 130 brain MRI images were selected from adult patients without pathological findings, ensuring assessment of fundamental sequence recognition rather than diagnostic capability. Images represented 13 standard MRI series, including axial T1-weighted, axial T2-weighted, axial FLAIR, coronal FLAIR, sagittal FLAIR, coronal T2-weighted, sagittal T1-weighted, axial SWI, axial DWI, axial ADC, and contrast-enhanced variants in multiple planes.
Data Acquisition and Anonymization: Images were obtained from hospital PACS (Picture Archiving and Communication System) using 1.5 Tesla scanners from Siemens and GE with similar sequence protocols. All images were thoroughly anonymized, exported in high-quality JPEG format (minimum resolution 994 × 1382 pixels) without compression, cropping, or visual post-processing.
Evaluation Protocol: Each image was individually uploaded to model interfaces using a standardized zero-shot prompt in English. To prevent in-context adaptation bias, a new session was initiated for each query by clearing chat history. All evaluations were conducted between June 23-29, 2025, using the most up-to-date model versions available.
Statistical Analysis: Model performance was evaluated based on accuracy calculations. Cochran's Q test and pairwise McNemar tests with Bonferroni correction were applied for the primary outcome (MRI sequence classification). Additional metrics included macro-averaged F1 scores, Cohen's kappa coefficients, and bootstrap resampling with 1000 iterations for stability estimates.
This methodological framework establishes a reproducible standard for evaluating computational optimization approaches in clinical MRI classification tasks.
Comparative Analysis of Optimization Approaches: RAG vs. Local Fine-Tuning
Technical Architecture and Performance Characteristics
Retrieval-Augmented Generation (RAG) and local fine-tuning represent distinct technical approaches to enhancing MLLM performance for clinical applications. RAG operates by decoupling knowledge storage from model parameters, using external knowledge bases that can be updated dynamically without model retraining [59]. In contrast, fine-tuning involves continuing the training of a pre-trained model on a specialized dataset, directly adjusting the model's internal weights to improve performance on specific tasks or domains [60] [59].
Table 2: Architectural Comparison of RAG vs. Fine-Tuning for Clinical MRI Applications
| Aspect | Retrieval-Augmented Generation (RAG) | Local Fine-Tuning |
|---|---|---|
| Supported Data | Dynamic, real-time updates | Static knowledge at training time |
| Setup Cost | Low (index configuration) | High (training resources) |
| Scalability | High, real-time knowledge updates | Low, requires model retraining |
| Update Time | Minutes | Hours/Days |
| Precision with Recent Changes | High (semantic search from updated sources) | Low unless retrained with new data |
| Data Privacy | Knowledge remains external, easier to control | Model internalizes knowledge, requiring secure training |
| Computational Requirements | Lower (leverages existing model) | Higher (requires training resources) |
| Interpretability | Source citation possible | Black-box decisions |
| Domain Adaptation | Controlled through knowledge base curation | Deep specialization through weight adjustment |
Performance Optimization and Limitations
Each approach presents distinct advantages and limitations for clinical deployment scenarios:
RAG Advantages and Implementation Considerations:
- Dynamic Knowledge Integration: RAG systems can incorporate the latest clinical guidelines, research findings, and institutional protocols by simply updating the knowledge base, avoiding the costly retraining cycle required by fine-tuning [59]. This is particularly valuable in rapidly evolving domains like neuro-oncology imaging.
- Enhanced Factual Accuracy and Traceability: By retrieving and citing specific source documents, RAG systems provide verifiable grounding for their outputs, reducing hallucinations and increasing clinical trust [61]. In one assessment, RAG provided correct answers with citable sources where fine-tuning produced inconsistent responses [62].
- Implementation Complexity: RAG introduces additional architectural components including retrievers, vector databases, and prompt engineering layers that require careful tuning [59]. Retrieval quality directly impacts system performance—irrelevant retrieved context can degrade response quality despite capable generators.
Fine-Tuning Advantages and Implementation Considerations:
- Domain-Specific Optimization: Fine-tuning enables deep specialization for specific clinical tasks. Techniques like Clinical Visual Instruction Tuning (CVIT) have demonstrated significant improvements in radiology report generation, with structured clinical templates and keyword-focused instructions enhancing report quality [7].
- Computational Efficiency Techniques: Parameter-efficient fine-tuning (PEFT) methods like LoRA (Low-Rank Adaptation) and QLoRA (Quantized LoRA) have dramatically reduced the computational barrier to fine-tuning [60]. LoRA adds small trainable matrices to model layers while freezing original weights, enabling efficient adaptation of large models on limited hardware.
- Catastrophic Forgetting Risk: Full fine-tuning can cause models to lose general capabilities while specializing, potentially reducing performance on tasks outside their fine-tuned domain [60]. This necessitates careful dataset design and evaluation.
Implementation Frameworks: Technical Protocols for Clinical Deployment
RAG Implementation Workflow for MRI Sequence Classification
The RAG architecture implements a multi-stage pipeline that enhances base model capabilities with domain-specific knowledge [61]:
Diagram 1: RAG Clinical Deployment Workflow
The RAG implementation process involves several critical phases:
Knowledge Base Curation: Clinical documentation including MRI protocol manuals, sequence characterization guidelines, and institutional imaging standards are collected. In a representative implementation, documents are converted to PDF format and contain structured question-answer pairs about domain-specific information [62].
Vectorization and Indexing: Documents are processed through chunking strategies optimized for clinical content, then converted to vector embeddings using clinical-domain models. These embeddings are indexed in specialized vector databases like Elasticsearch with semantic search capabilities [62].
Query Processing and Augmentation: User queries for MRI sequence identification are converted to embeddings, and semantic search retrieves the most relevant document chunks. The retrieved context is formatted into an augmented prompt that provides clinical grounding.
Generation and Verification: The base LLM generates responses incorporating both its inherent capabilities and the retrieved clinical context. Source citations enable clinical validation, with implementation showing correct answer generation with verifiable sources [62].
Local Fine-Tuning Protocol for Clinical MRI Specialization
Local fine-tuning adapts base models to clinical domains using institutional data while maintaining data privacy through on-premises deployment [60]:
Diagram 2: Clinical Fine-Tuning Protocol
The fine-tuning methodology encompasses several research-validated stages:
Clinical Dataset Curation: The 3D-BrainCT dataset exemplifies appropriate clinical data collection, containing 18,885 text-scan pairs with comprehensive lesion details including degree, spatial landmarks, and diagnostic impressions of both neuronal and vascular CT features [7]. Such datasets provide the foundation for effective clinical adaptation.
Instruction Tuning Methodologies: Research demonstrates that structured clinical instruction tuning significantly enhances model performance. The BrainGPT implementation compared four approaches: (1) Plain Instruction (stating the model's role as a radiology assistant), (2) In-Context Example Instruction (adding 3-shot examples), (3) Template Instruction (incorporating structured clinical QA templates), and (4) Keyword Instruction (providing categorical guidelines focused on keywords) [7]. The keyword instruction approach demonstrated superior performance in clinical essence metrics.
Parameter-Efficient Fine-Tuning: For computational optimization, LoRA (Low-Rank Adaptation) introduces small trainable matrices to model layers while freezing original weights, dramatically reducing memory requirements [60]. QLoRA extends this approach by quantizing the base model to 4-bit precision, enabling fine-tuning of large models on single GPUs [60].
Clinical Evaluation Framework: Traditional NLP metrics like BLEU and ROUGE show poor correlation with clinical utility. The FORTE (Feature-Oriented Radiology Task Evaluation) framework addresses this by evaluating four essential components: degree, landmark, feature, and impression [7]. In validation studies, BrainGPT achieved an average FORTE F1-score of 0.71, with 74% of generated reports being indistinguishable from human-written ground truth in Turing-like tests [7].
Integrated Deployment Strategy: A Hybrid Approach for Clinical Applications
Combining RAG and Fine-Tuning for Optimized Clinical Performance
A hybrid approach that strategically combines local fine-tuning with RAG augmentation delivers superior performance for clinical MRI applications while addressing privacy and computational constraints [62]. This integrated methodology leverages the complementary strengths of both paradigms:
Domain-Specialized Foundation: Begin with a clinically fine-tuned base model (such as BrainGPT for neuroimaging) that encapsulates fundamental medical knowledge and terminology. Fine-tuning with clinical visual instruction tuning (CVIT) establishes baseline competence in radiological reasoning and reporting conventions [7].
Dynamic Knowledge Augmentation: Layer RAG capabilities atop the fine-tuned model to incorporate institution-specific protocols, latest research findings, and evolving clinical guidelines. This combination proved effective in implementations where domain-specific fine-tuning established foundational knowledge, while RAG provided current, verifiable information [62].
Privacy-Preserving Architecture: Deploy the hybrid system on-premises using Kubernetes-based workflows or high-end hardware like NVIDIA DGX systems, ensuring patient data never leaves institutional control [60]. Federated learning approaches can further enhance privacy by enabling model refinement across institutions without centralizing sensitive data [60].
Implementation Tools and Research Reagents
Successful deployment requires a carefully selected toolkit of frameworks and platforms that support both RAG and fine-tuning capabilities:
Table 3: Essential Research Reagents and Computational Tools for Clinical MLLM Deployment
| Tool Category | Representative Solutions | Clinical Application Function |
|---|---|---|
| RAG Orchestration | LangChain, LlamaIndex, Haystack | Manages document ingestion, vectorization, retrieval, and generation pipelines for clinical QA systems |
| Vector Databases | Pinecone, Weaviate, Qdrant, FAISS | Stores and retrieves clinical document embeddings with high precision and low latency |
| Evaluation Frameworks | Ragas, FORTE (Feature-Oriented Radiology Task Evaluation) | Measures clinical accuracy beyond traditional NLP metrics, focusing on diagnostic relevance |
| Fine-Tuning Frameworks | PEFT (Parameter-Efficient Fine-Tuning), LoRA, QLoRA | Enables computational-efficient model adaptation to clinical domains with limited data |
| On-Premises Deployment | NVIDIA DGX Systems, Kubernetes with Kubeflow, Ray on Anyscale | Provides secure, scalable infrastructure for privacy-compliant clinical model deployment |
| Specialized Clinical Models | BrainGPT (3D CT), Med-PaLM Multimodal, LLaVA-Med | Domain-adapted foundation models pre-trained on medical data for clinical applications |
The integration of RAG and local fine-tuning technologies presents a compelling pathway for deploying multimodal LLMs in clinical brain MRI applications while addressing critical computational and privacy concerns. Performance benchmarks demonstrate that while general-purpose MLLMs show promise in MRI sequence classification, significant performance gaps remain that require domain-specific optimization.
RAG architectures provide dynamic knowledge integration with enhanced traceability, making them ideal for incorporating the latest clinical evidence and institutional protocols. Local fine-tuning enables deep domain specialization through clinical visual instruction tuning and parameter-efficient adaptation methods. A hybrid approach that combines a fine-tuned clinical foundation model with RAG-based dynamic knowledge retrieval offers the most promising framework for clinical deployment.
Implementation should prioritize privacy-preserving on-premises architectures, clinical-grade evaluation using domain-specific metrics like FORTE, and careful integration with existing clinical workflows. As multimodal LLMs continue to evolve, these computational and privacy optimization strategies will play an increasingly vital role in translating AI capabilities into clinically valuable tools that enhance diagnostic accuracy while safeguarding patient data.
Benchmarking MLLM Performance: A Comparative Analysis of Leading Models in Brain MRI Tasks
The integration of Multimodal Large Language Models (LMMs) into brain MRI analysis represents a paradigm shift in medical imaging research. These models are demonstrating remarkable capabilities in processing and interpreting complex neuroimaging data. This comparative framework objectively evaluates the performance of leading LMMs across three critical tasks in brain MRI research: sequence classification, Visual Question Answering (VQA), and medical report generation. By synthesizing experimental data and methodologies from recent studies, this guide provides researchers, scientists, and drug development professionals with actionable insights for model selection and implementation in neuroscientific discovery and clinical applications.
Performance Comparison Tables
Model Performance Across Brain MRI Tasks
Table 1: Performance comparison of specialized models and LMMs on specific brain MRI tasks.
| Task | Model Name | Architecture | Dataset | Performance Metrics |
|---|---|---|---|---|
| Sequence Classification | GPT-4-based LLM [12] | Not Specified | 1,490 brain MRI sequences (UCSF) | Accuracy: 0.83 (Outperformed CNN & string-matching) |
| MedViT (Benchmark) [11] | CNN-Transformer Hybrid | Pediatric CNS Tumors (2,383 sequences) | Accuracy: 0.893 (95% CI 0.880–0.904) | |
| MedViT (Expert Adjusted) [11] | CNN-Transformer Hybrid | Pediatric CNS Tumors (2,383 sequences) | Accuracy: 0.905 (95% CI 0.893–0.916) | |
| Visual Question Answering (VQA) | mpLLM [29] [63] | Prompt-conditioned Hierarchical MoE | Multi-parametric 3D Brain MRI | Outperformed strong medical VLM baselines by +5.3% on average |
| Medical Report Generation | MRG-LLM [64] | Frozen LLM + Learnable Visual Encoder | IU X-ray, MIMIC-CXR | Achieved state-of-the-art performance |
Table 2: Key specifications of leading general-purpose Multimodal LMMs (2025).
| Model Name | Developer | Key Capabilities | Context Window | License |
|---|---|---|---|---|
| Gemma 3 [20] [65] | Google DeepMind | Vision-Language input, text output, dynamic image processing | 128K tokens | Open weights, responsible commercial use |
| Qwen 2.5 VL [20] [65] | Alibaba Cloud | Object recognition, scene interpretation, multilingual support | 32K tokens (extensible with YaRN) | Apache 2.0 |
| Pixtral Large [20] | Mistral AI | Integrates visual and textual data, function calls | 128K tokens | Mistral Research License |
| Phi-4 Multimodal [20] | Microsoft | Unified vision, audio, and text processing, low-latency | 128K tokens | MIT |
| Llama 3.2 Vision [20] | Meta | Multimodal reasoning, optimized for various hardware | 128K tokens | Community License (specific terms) |
Experimental Protocols and Methodologies
MRI Sequence Classification
3.1.1 Protocol for LLM-based Classification (GPT-4) The study evaluated a GPT-4-based classifier on 1,490 brain MRI sequences from UCSF, comparing its performance against traditional Convolutional Neural Networks (CNNs) and string-matching methods. The primary metrics were sensitivity, specificity, and accuracy. The LLM classifier demonstrated superior performance, achieving an accuracy of 0.83. A key advantage noted was the model's interpretability, which provided additional insights and improved classification transparency, thereby minimizing false positives [12].
3.1.2 Protocol for Handling Domain Shift (MedViT) This research addressed the critical challenge of domain shift, particularly when applying models trained on adult data to pediatric MRI datasets. The methodology involved [11]:
- Model Architecture Comparison: A pre-trained ResNet-18 (CNN) was compared against MedViT, a hybrid CNN-Transformer model, on a pediatric dataset containing 2,383 sequences (T1, T2, CT1, FLAIR, SWI, T2*).
- Expert Domain Knowledge Integration: Model performance was enhanced by adjusting its decision-making process to ignore labels absent from the pediatric test set, aligning the classification task with the new data's characteristics.
- Results: The hybrid MedViT architecture (accuracy 0.893) demonstrated superior robustness to domain shift compared to ResNet-18. Expert adjustment further improved MedViT's accuracy to 0.905.
Visual Question Answering on 3D Brain MRI
Protocol for mpLLM The mpLLM study introduced a novel approach for Visual Question Answering on multi-parametric 3D Brain MRI (mpMRI). The methodology had several key components [29] [63]:
- Model Architecture: A prompt-conditioned hierarchical Mixture-of-Experts (MoE) that routes across modality-level and token-level projection experts. This design fuses multiple interrelated 3D modalities efficiently without requiring image-report pretraining.
- Synthetic VQA Protocol: To overcome the scarcity of image-text paired data, the model integrated a synthetic VQA protocol that generated medically relevant questions and answers from existing segmentation annotations.
- Clinical Validation: The generated VQA dataset was clinically validated in collaboration with medical experts, ensuring its relevance and accuracy.
- Performance: mpLLM outperformed strong medical VLM baselines by an average of 5.3% across multiple mpMRI datasets.
Medical Report Generation
Protocol for MRG-LLM The MRG-LLM framework was designed for generating medical reports from imaging data. Its key innovation lies in a dynamic prompt customization mechanism [64]:
- Architecture: MRG-LLM combines a frozen LLM with a learnable visual encoder.
- Dynamic Prompt Customization: The core of the method generates instance-specific prompts tailored to individual medical images. This is achieved through conditional affine transformations derived from the image's visual features.
- Implementation: The study proposed two implementations of this mechanism: prompt-wise and promptbook-wise customization, enabling precise and targeted report generation.
- Evaluation: Extensive experiments on standard public datasets like IU X-ray and MIMIC-CXR demonstrated that MRG-LLM achieves state-of-the-art performance.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential datasets, models, and computational resources for brain MRI LMM research.
| Resource Name | Type | Primary Function | Relevance to Brain MRI Research |
|---|---|---|---|
| MICBench (Multiple Images Comparison Benchmark) [66] | Dataset | Benchmark for multi-image quality comparison tasks | Provides 4,000 human-annotated MCQs for evaluating LMMs on visual quality reasoning. |
| Co-Instruct-562K [66] | Dataset | Large-scale instruction-tuning dataset for visual quality comparison | Enables training of LMMs for fine-grained, open-ended quality assessment. |
| MedViT [11] | Model | CNN-Transformer hybrid for medical image classification | Proven effective for MRI sequence classification, especially under domain shift. |
| vLLM / Ollama [20] | Computational Framework | High-throughput inference and local deployment of LLMs | Facilitates scalable deployment and serving of open-source LMMs. |
| Serverless GPUs (e.g., Koyeb) [20] | Computational Resource | Scalable, no-infrastructure GPU computing | Enables cost-effective fine-tuning and deployment of large models without managing complex infrastructure. |
This comparative framework synthesizes the current state of Multimodal LLMs applied to brain MRI research, highlighting specialized architectures like mpLLM for VQA and MRG-LLM for report generation, alongside the robust performance of general-purpose models like GPT-4 in classification. The consistent theme across studies is that success hinges on tailored architectures—such as MoEs for multi-parametric data and hybrid CNN-Transformers for handling domain shift—coupled with strategies to overcome data scarcity. For researchers in neuroscience and drug development, the choice of model is task-specific: MedViT excels in cross-domain sequence classification, mpLLM is pioneering for 3D MRI VQA, and MRG-LLM sets a new standard for automated report generation. As the field evolves, the integration of these tools with clinical expertise and scalable computational resources will be crucial for translating multimodal AI into impactful biomedical discoveries.
Multimodal large language models (MLLMs) represent a transformative advancement in artificial intelligence, with significant implications for visually intensive disciplines such as medical imaging. Within radiology, the accurate interpretation of brain magnetic resonance imaging (MRI) requires precise identification of different sequence types, as each sequence provides unique contrast mechanisms to highlight specific tissue characteristics and pathological findings. This comparative analysis examines the capabilities of three leading MLLMs—ChatGPT-4o (OpenAI), Gemini 2.5 Pro (Google), and Claude 4 Opus (Anthropic)—in classifying brain MRI sequences, a fundamental task that underpins more complex diagnostic applications [9] [1]. Understanding the relative strengths and limitations of these models provides crucial insights for researchers and clinicians considering their integration into brain imaging analysis workflows.
Experimental Protocols and Methodologies
Dataset Composition and Image Selection
The foundational study comparing these three models utilized a carefully curated dataset of 130 brain MRI images acquired from adult patients without pathological findings [9] [1]. This dataset comprehensively represented 13 standard MRI series essential in clinical neuroradiology:
- Basic structural sequences: Axial T1-weighted (T1w), axial T2-weighted (T2w), sagittal T1w
- Fluid-sensitive sequences: Axial fluid-attenuated inversion recovery (FLAIR), coronal FLAIR, sagittal FLAIR, coronal T2w
- Advanced and functional sequences: Axial susceptibility-weighted imaging (SWI), axial diffusion-weighted imaging (DWI), axial apparent diffusion coefficient (ADC)
- Contrast-enhanced sequences: Contrast-enhanced axial T1w, contrast-enhanced coronal T1w, contrast-enhanced sagittal T1w
All images were obtained from 1.5 Tesla scanners from two major manufacturers (Siemens Healthcare and GE Healthcare) to represent equipment variability encountered in clinical practice. To ensure standardization, a single representative slice was selected for each series at an anatomical level where the lateral ventricles were clearly visible. Images were exported in high-quality JPEG format (minimum resolution: 994 × 1382 pixels) without compression, cropping, or visual post-processing, and contained no annotations, arrows, or textual markings that could influence model interpretation [9].
Evaluation Framework and Prompt Design
The study employed a zero-shot prompting approach where each model processed individual images without prior examples or fine-tuning. The standardized prompt explicitly stated the research context and absence of clinical application to mitigate potential response biases [9]:
"This is a medical research question for evaluation purposes only. Your response will not be used for clinical decision-making. No medical responsibility is implied. Please examine this medical image and answer the following questions: What type of radiological modality is this examination? Which anatomical region does this examination cover? What is the imaging plane (axial, sagittal, or coronal)? Is this a contrast-enhanced image or not? If this image is an MRI, what is the specific MRI sequence? If it is not an MRI, write 'Not applicable.' Please number your answers clearly."
To prevent in-context adaptation, where models might alter responses based on previous interactions, a new session was initiated for each prompt by clearing the chat history. All evaluations were conducted between June 23, 2025, and June 29, 2025, using the most up-to-date versions of each model available at that time. Model responses were independently reviewed and classified as "correct" or "incorrect" by two radiologists working in consensus [9].
Statistical Analysis
The primary outcome measure was accuracy in MRI sequence classification. Statistical comparisons between models for this task utilized Cochran's Q test for overall performance differences, followed by pairwise McNemar tests with Bonferroni correction for specific model-to-model comparisons. In addition to accuracy, the study calculated macro-averaged F1 scores and Cohen's kappa coefficients to evaluate inter-class performance consistency and agreement with ground truth. Bootstrap resampling with 1000 iterations provided 95% confidence intervals for sequence-specific accuracy estimates, addressing concerns related to the limited number of images per sequence class [9] [67].
MRI Sequence Classification Study Workflow
Performance Metrics and Comparative Analysis
Core Recognition Task Performance
All three models demonstrated perfect or near-perfect performance in basic image recognition tasks, achieving 100% accuracy in identifying the imaging modality (MRI) and anatomical region (brain). ChatGPT-4o and Gemini 2.5 Pro maintained perfect 100% accuracy in imaging plane classification (axial, sagittal, coronal), while Claude 4 Opus achieved 99.23% accuracy (129/130) in this task. In assessing contrast-enhancement status, both ChatGPT-4o and Gemini 2.5 Pro achieved 98.46% accuracy (128/130), with Claude 4 Opus slightly lower at 95.38% (124/130) [9].
The most significant performance differentiator emerged in the central task of specific MRI sequence classification, where models demonstrated substantially varied capabilities as shown in Table 1.
Table 1: Comparative Performance Across MRI Analysis Tasks
| Classification Task | ChatGPT-4o | Gemini 2.5 Pro | Claude 4 Opus |
|---|---|---|---|
| Modality Identification | 100% | 100% | 100% |
| Anatomical Region Recognition | 100% | 100% | 100% |
| Imaging Plane Classification | 100% | 100% | 99.23% |
| Contrast-Enhancement Status | 98.46% | 98.46% | 95.38% |
| MRI Sequence Classification | 97.69% | 93.08% | 73.08% |
MRI Sequence Classification Details
The substantial performance differences in MRI sequence classification warranted deeper analysis of error patterns and model-specific limitations. ChatGPT-4o achieved the highest accuracy at 97.69% (127/130), misclassifying only three images. Gemini 2.5 Pro demonstrated strong but comparatively lower performance at 93.08% (121/130), while Claude 4 Opus showed significantly reduced accuracy at 73.08% (95/130) [9] [1].
Error analysis revealed that the most frequent misclassifications across models involved fluid-attenuated inversion recovery (FLAIR) sequences, which were often incorrectly identified as T1-weighted or diffusion-weighted sequences. Claude 4 Opus demonstrated particular difficulties with susceptibility-weighted imaging (SWI) and apparent diffusion coefficient (ADC) sequences, suggesting specific gaps in its training data or architectural limitations for these specialized sequences [9].
A notable finding was Gemini 2.5 Pro's tendency to occasionally produce hallucinations, including irrelevant clinical details such as "hypoglycemia" and "Susac syndrome" in its responses. While these hallucinations did not always directly impact sequence classification accuracy, they raise important concerns about clinical reliability and the potential for misleading outputs in real-world applications [9] [1].
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for LLM Evaluation in Medical Imaging
| Research Component | Specification | Function in Experimental Design |
|---|---|---|
| Brain MRI Dataset | 130 images, 13 sequences, normal findings | Provides standardized testbed for sequence recognition capability assessment |
| Clinical MRI Scanners | 1.5 Tesla (Siemens MAGNETOM, GE Optima 360) | Ensures representative image quality and clinical relevance |
| Zero-shot Prompt Framework | Standardized English text with research disclaimer | Controls for prompt variability and enables reproducible model comparisons |
| Radiologist Ground Truth | Two radiologists in consensus | Establishes reference standard for model performance evaluation |
| Statistical Analysis Package | SPSS version 28.0 with bootstrap resampling | Enforms rigorous statistical comparisons and confidence interval estimation |
Implications for Brain MRI Analysis Research
The performance differentials observed in this comparative analysis have significant implications for research applications in brain MRI analysis. ChatGPT-4o's superior performance in sequence classification (97.69%) positions it as the current leading candidate for applications requiring reliable sequence identification, such as automated image sorting or quality control pipelines [9] [1].
However, the observed limitations in more complex diagnostic tasks highlighted in complementary research temper enthusiasm for immediate clinical deployment. A separate study evaluating ChatGPT-4o in brain tumor diagnosis found that while the model achieved high accuracy in identifying basic MRI features (88.3% for sequences, 81% for perilesional edema, 79.5% for signal characteristics), it significantly underperformed in diagnostic reasoning tasks. Specifically, ChatGPT-4o achieved only 56.8% accuracy for differential diagnoses and 29.5% for most likely diagnoses compared to 93.2-90.9% and 70.5-65.9% for human radiologists, respectively [68] [69].
The "expertise paradox" identified in LLM-assisted diagnostic workflows further refines our understanding of optimal integration strategies. Research demonstrates that LLM-generated differential diagnoses achieve highest accuracy when provided with image descriptions from neuroradiologists (top-3 accuracy: 78.8-83.8%), followed by radiology residents (71.8-77.6%), and finally neurology/neurosurgery residents (62.6-64.5%). Paradoxically, relative diagnostic gains through LLM assistance diminish with increasing reader expertise, with neurology/neurosurgery residents showing +19.2% improvement compared to only +4.4% for neuroradiologists [70].
This comprehensive comparison reveals a rapidly evolving but maturing landscape for multimodal LLMs in brain MRI sequence classification. ChatGPT-4o currently establishes the performance benchmark at 97.69% accuracy, followed closely by Gemini 2.5 Pro at 93.08%, with Claude 4 Opus significantly trailing at 73.08%. These quantitative performance metrics provide crucial guidance for researchers selecting models for neuroimaging applications.
The consistent observation of hallucinations across models, particularly with Gemini 2.5 Pro, alongside persistent challenges in complex diagnostic reasoning tasks, underscores that human expertise remains indispensable in clinical settings. Future research directions should prioritize hallucination mitigation, specialized training on less common MRI sequences, and optimized human-AI collaboration frameworks that leverage the complementary strengths of both computational and human intelligence in brain MRI analysis.
The adoption of multimodal large language models (MLLMs) in clinical brain MRI analysis presents a critical evaluation challenge. While these models demonstrate remarkable capabilities in classifying MRI sequences and generating diagnostic reports, traditional natural language processing (NLP) metrics fail to capture their true clinical utility. Research reveals that standard metrics like BLEU, METEOR, and ROUGE-L, designed for machine translation and text summarization, are inherently insensitive to the clinical essence of generated radiology reports [7]. These metrics primarily measure superficial text similarity through n-gram overlap or longest common subsequences, but cannot assess whether critical pathological findings are correctly identified, localized, and described with appropriate clinical terminology [28].
This evaluation gap becomes particularly problematic in brain MRI sequence classification, where precise identification of imaging sequences and accurate feature description are fundamental to diagnostic accuracy. Recent studies evaluating multimodal LLMs like ChatGPT-4o, Claude 4 Opus, and Gemini 2.5 Pro on brain MRI recognition tasks demonstrate that while these models achieve high accuracy on basic recognition tasks (98.46-100% for modality, plane, and contrast status identification), their performance varies significantly on specific sequence classification (73.08-97.69%) [9]. More concerningly, these models occasionally exhibit hallucinations, generating clinically irrelevant details that could mislead diagnostic decisions [9]. These findings underscore the critical need for an evaluation framework that specifically addresses clinical relevance and accuracy.
FORTE: A Framework for Feature-Oriented Radiology Task Evaluation
Conceptual Foundation and Design Principles
The Feature-Oriented Radiology Task Evaluation (FORTE) framework represents a paradigm shift in assessing medical MLLM outputs. Developed specifically to address the limitations of traditional metrics, FORTE introduces a structured keyword extraction concept that captures the multi-semantic context of radiology reports [7]. This framework is designed with three core principles: (1) addressing the multi-semantic context of radiology reports, (2) recognizing synonyms to detect broader arrays of related terms, and (3) ensuring transferability across multiple imaging modalities [7].
FORTE operates by categorizing radiology keywords into four essential components that reflect the logical structure of clinical reasoning in neuroradiology. The "Degree" component evaluates how well the model indicates intensity or state (e.g., normal, mild, chronic, acute). The "Landmark" component assesses precise spatial localization (e.g., intracerebral, periventricular, midline). The "Feature" component focuses on accurate description of abnormalities (e.g., hemorrhage, atrophy, infarcts, mass effect). Finally, the "Impression" component evaluates clinical synthesis and diagnostic conclusions (e.g., arteriosclerotic encephalopathy, intracerebral hemorrhage) [7] [28].
Implementation and Scoring Methodology
FORTE implements a systematic approach to extract and categorize clinical concepts from generated reports and compare them against ground truth annotations. The framework employs a synonym-aware matching system that recognizes equivalent clinical terms, thereby addressing the variability in clinical language. For each category—Degree, Landmark, Feature, and Impression—precision, recall, and F1-scores are calculated, providing a multi-faceted assessment of clinical information capture [7].
The final FORTE score represents a composite measure that reflects the model's ability to capture clinically essential information across all categories. In validation studies, the framework has demonstrated high sensitivity to improvements in clinical instruction tuning, with BrainGPT models achieving an average FORTE F1-score of 0.71 (degree = 0.661; landmark = 0.706; feature = 0.693, and impression = 0.779) [7]. Notably, 74% of reports generated by the FORTE-optimized BrainGPT model were indistinguishable from human-written ground truth in Turing-like tests conducted by physician raters [7].
FORTE Evaluation Workflow for Brain MRI Analysis
Comparative Performance Analysis: FORTE vs. Traditional Metrics
Quantitative Comparison Across Evaluation Frameworks
The table below summarizes the performance disparities between traditional metrics and FORTE when evaluating MLLM-generated brain imaging reports:
| Evaluation Metric | Core Mechanism | Sensitivity to Clinical Relevance | Optimal Use Case | BrainGPT Performance |
|---|---|---|---|---|
| FORTE (F1-Score) | Structured keyword extraction across 4 clinical categories | High | Clinical report generation | 0.710 (average) |
| CIDEr-R | TF-IDF weighted n-gram similarity | Moderate | General medical image captioning | 153.3 (after sentence pairing) |
| BLEU-4 | N-gram overlap precision | Low | Machine translation | ~0 (baseline), minimal improvement after tuning |
| METEOR | Word-to-word matching with synonyms | Low | Machine translation | Minimal improvement with advanced tuning |
| ROUGE-L | Longest common subsequence | Low | Text summarization | Moderate improvement after sentence pairing |
This comparison reveals fundamental limitations in traditional metrics. Notably, BLEU-4 scored approximately zero for baseline models, indicating negligible n-gram overlap despite clinically relevant outputs [7]. While CIDEr-R showed some sensitivity to clinical keyword usage through its TF-IDF component, it still failed to capture the structural clinical reasoning embodied in FORTE's categorical framework [7] [28].
Multimodal LLM Performance on Brain MRI Sequence Classification
Recent comprehensive evaluations of state-of-the-art multimodal LLMs on brain MRI sequence classification tasks provide critical context for FORTE's application:
| Multimodal LLM | MRI Sequence Classification Accuracy | Contrast-Enhancement Status Accuracy | Imaging Plane Classification Accuracy | Key Limitations |
|---|---|---|---|---|
| ChatGPT-4o | 97.69% (127/130) | 98.46% (128/130) | 100% (130/130) | Occasional misclassification of FLAIR sequences |
| Gemini 2.5 Pro | 93.08% (121/130) | 98.46% (128/130) | 100% (130/130) | Hallucinations (e.g., "hypoglycemia", "Susac syndrome") |
| Claude 4 Opus | 73.08% (95/130) | 95.38% (124/130) | 99.23% (129/130) | Lower accuracy on SWI and ADC sequences |
| BrainGPT (3D CT) | N/A (CT-focused) | N/A (CT-focused) | N/A (CT-focused) | Specialized for 3D CT report generation |
This evaluation, conducted on 130 brain MRI images across 13 standard sequences, demonstrates that while multimodal LLMs excel at basic recognition tasks, their performance varies significantly on clinically critical sequence discrimination [9]. The most frequent misclassifications involved fluid-attenuated inversion recovery (FLAIR) sequences, often misidentified as T1-weighted or diffusion-weighted sequences [9]. These findings highlight the need for FORTE-like frameworks that can detect such clinically significant errors that might be overlooked by conventional accuracy metrics alone.
Experimental Protocols and Methodologies
BrainGPT Development and Training Protocol
The development of BrainGPT, which pioneered the FORTE evaluation approach, followed a rigorous experimental protocol centered on clinical visual instruction tuning (CVIT). The model was built upon the open-source Otter framework, which utilizes OpenFlamingo's architecture with a CLIP ViT-L/14 visual encoder, Perceiver Resampler, and LLaMA-7B language model [28]. The training incorporated 18,885 text-scan pairs from the 3D-BrainCT dataset, comprising brain CT scans from Alzheimer's patients with an average age exceeding 82 years [7] [28].
The experimental design compared four distinct instruction-tuning conditions: (1) Plain Instruction (basic role definition as radiology assistant), (2) In-context Example Instruction (adding 3-shot examples), (3) Template Instruction (incorporating structured clinical QA templates), and (4) Keyword Instruction (providing categorical guidelines focusing on degree, landmark, feature, and impression) [7]. This progressive refinement of clinical context enabled the systematic improvement of clinical reporting quality, which FORTE subsequently quantified through its structured evaluation approach.
Multimodal LLM Evaluation Protocol for Brain MRI Classification
The comparative analysis of multimodal LLMs followed a standardized zero-shot prompting protocol. Researchers utilized 130 brain MRI images from adult patients without pathological findings, representing 13 standard MRI sequences [9]. Each model received identical prompts requesting classification of: (1) radiological modality, (2) anatomical region, (3) imaging plane, (4) contrast-enhancement status, and (5) specific MRI sequence [9].
To prevent in-context adaptation bias, a new session was initiated for each prompt by clearing chat history [9]. All evaluations were conducted between June 23-29, 2025, using the most up-to-date model versions available at that time. Responses were independently reviewed and classified as "correct" or "incorrect" by two radiologists in consensus, with hallucinations defined as statements unrelated to the input image or prompt context [9].
Sentence Pairing Enhancement Technique
A crucial methodological innovation that bridged traditional metrics and clinical evaluation was sentence pairing. Recognizing that traditional metrics struggle with paragraph-structured reports, researchers decomposed multi-sentence paragraphs into smaller semantic units through cosine similarity-based pairing [7] [28]. This process significantly enhanced metric scores (average increase of 5.28 points in METEOR, 6.48 in ROUGE-L, and 114 points in CIDEr-R) while maintaining clinical relevance [7]. The technique particularly benefited advanced CVIT models, with CIDEr-R scores showing a prominent ascending trend across the instruction-tuning hierarchy: BrainGPT-plain (125.86), BrainGPT-example (132.38), BrainGPT-template (147.92), and BrainGPT-keyword (153.3) [7].
BrainGPT Experimental Workflow with FORTE Validation
| Resource Category | Specific Tools/Datasets | Function in Evaluation | Key Characteristics |
|---|---|---|---|
| Medical Imaging Datasets | 3D-BrainCT Dataset (18,885 text-scan pairs) | Training and validation of MLLMs for brain imaging | 3D CT scans from Alzheimer's patients (>82 years average age) [7] |
| Multimodal LLM Frameworks | Otter/OpenFlamingo (BrainGPT base) | Foundation architecture for medical MLLMs | CLIP ViT-L/14 encoder + Perceiver Resampler + LLaMA-7B [28] |
| Evaluation Metrics | FORTE Framework | Clinical essence assessment | Four-category keyword extraction (Degree, Landmark, Feature, Impression) [7] |
| Clinical Validation Tools | Physician Turing Test | Real-world clinical validation | 11 physician raters, 74% reports indistinguishable from human [7] |
| Preprocessing Techniques | Sentence Pairing | Enhanced metric evaluation | Cosine similarity-based sentence decomposition [7] |
| Benchmark LLMs | ChatGPT-4o, Gemini 2.5 Pro, Claude 4 Opus | Comparative performance benchmarking | Standardized zero-shot evaluation on 130 brain MRI images [9] |
Implications for Brain MRI Sequence Classification Research
The FORTE framework establishes a crucial foundation for advancing brain MRI sequence classification research by addressing three critical challenges. First, it enables precise quantification of clinical information retention in generated reports, moving beyond simple sequence identification to assess how comprehensively findings are communicated [7] [9]. Second, FORTE's structured approach facilitates targeted model improvement by identifying specific clinical categories (degree, landmark, feature, or impression) where models underperform [7]. Third, the framework supports cross-modal evaluation consistency, allowing comparable assessment across different imaging technologies (CT vs. MRI) and clinical domains [7].
Recent findings from multimodal LLM evaluations further underscore FORTE's relevance. The observed hallucinations in models like Gemini 2.5 Pro (generating irrelevant clinical details such as "hypoglycemia" and "Susac syndrome") [9] represent precisely the type of clinical safety issue that traditional metrics would miss but FORTE can systematically detect through its feature-oriented analysis. Similarly, the variable performance across MRI sequences (particularly challenges with FLAIR, SWI, and ADC sequences) [9] highlights the need for evaluation frameworks that can discriminate between clinically significant and insignificant classification errors.
The integration of FORTE into the broader AI assessment ecosystem, complementing frameworks like AI for IMPACTS (which evaluates integration, monitoring, performance, acceptability, cost, technological safety, and scalability) [71], represents a necessary evolution toward comprehensive medical AI validation. This multi-dimensional approach ensures that models demonstrating technical proficiency in controlled evaluations also deliver tangible clinical benefits in real-world settings, ultimately accelerating the responsible integration of multimodal LLMs into clinical workflows for brain MRI analysis and beyond.
The accurate classification of brain MRI sequences represents a complex clinical task where the distinction between thinking (reasoning-intensive) and non-thinking (direct, automated) cognitive processes becomes critical. This comparison guide analyzes the performance of multimodal Large Language Models (LLMs) operating in these distinct modes within the specific context of brain MRI classification research. In clinical neuroscience, analytic thinking involves focused, detail-oriented processing localized to specific neural pathways, while holistic thinking employs broader, integrative pattern recognition across distributed brain networks [72]. Modern multimodal LLMs have begun to emulate these distinct processing approaches, offering researchers powerful tools to advance neurodiagnostic capabilities. The neural mechanisms distinguishing these thinking styles involve coordinated activity across the bilateral frontal and parietal lobes, precentral and postcentral gyri, and supplementary motor areas [72], creating a biological framework for understanding how AI systems might approach similar classification tasks.
Theoretical Framework: Thinking and Non-Thinking Modes
Defining Cognitive Modes in Humans and AI
In both human cognition and artificial intelligence, "thinking" and "non-thinking" modes represent distinct approaches to information processing:
Thinking Modes (Reasoning-Intensive): These approaches involve multi-step analysis, conscious deliberation, and logical inference chains. In humans, this corresponds to conscious reasoning with intent to reach conclusions using logically justified methods [73]. In AI, this manifests as models that engage in chain-of-thought reasoning, verification processes, and iterative analysis before generating outputs [74].
Non-Thinking Modes (Direct Processing): These approaches utilize automated, pattern-based responses with minimal conscious deliberation. In humans, this aligns with perceptuo-motor functions processed rapidly in hardwired, spatially segregated neural modules [73]. In AI, this corresponds to models that generate immediate, single-step responses without explicit reasoning steps.
Neural Basis of Thinking Styles
fMRI research reveals distinct neural correlates for different thinking approaches. Holistic thinking engages widespread bilateral networks including frontal lobes, parietal lobes, fusiform, and insula regions [72]. Conversely, analytic thinking shows more focused activation patterns with heightened engagement in regions supporting detailed feature analysis [72]. These biological insights inform the development of AI architectures that can simulate similar specialized processing for clinical tasks.
Table 1: Neural Correlates of Thinking Styles Relevant to Clinical MRI Classification
| Thinking Style | Key Brain Regions | Processing Characteristics | Clinical Classification Strengths |
|---|---|---|---|
| Analytic Thinking | Bilateral frontal lobes, precentral gyrus, supplementary motor area [72] | Focused, detail-oriented, sequential processing | Fine-grained lesion detection, subtle anomaly identification |
| Holistic Thinking | Bilateral parietal lobes, fusiform, insula, angular gyrus [72] | Integrative, pattern-based, contextual processing | Overall pattern recognition, multi-feature integration |
Experimental Approaches for Evaluating Reasoning Capabilities
fMRI Data Classification Methodologies
Research on brain activity classification provides established experimental protocols for evaluating thinking modes:
Task Paradigm Design: Studies use trial-based designs with specific thought tasks (e.g., motor imagery, mental calculation, visual imagery) performed over short time periods (<5 seconds) during fMRI acquisition [75]. This approach captures distinct neural signatures of different cognitive processes.
Feature Extraction: Regions of Interest (ROI)-based feature vectors are automatically extracted from activation maps. The selection focuses on regions consistently and exclusively activated for specific tasks during training processes [75].
Classification Algorithms: Support Vector Machine (SVM) algorithms with parameter optimization through k-fold cross-validation successfully identify thought tasks with mean accuracy of 74.5% (±14.3%) across subjects [75]. Modern approaches also employ convolutional neural networks and discriminant analysis [76].
Evaluating LLM Reasoning in Clinical Contexts
Multimodal LLMs with enhanced reasoning capabilities can be evaluated using adapted neuroscience protocols:
Stimulus Presentation: Clinical MRI images paired with diagnostic questions presented to LLMs in standardized formats.
Response Analysis: Comparison of outputs from models with "thinking" modes enabled versus standard direct generation.
Accuracy Validation: Expert radiologist assessment of diagnostic suggestions to establish ground truth references.
The following diagram illustrates a standardized experimental workflow for evaluating thinking versus non-thinking modes in clinical MRI classification tasks:
Comparative Analysis of Multimodal LLMs in Clinical Tasks
Leading Multimodal LLMs with Advanced Reasoning
Recent advancements in multimodal LLMs have produced several models with exceptional capabilities for clinical image analysis:
Table 2: Advanced Multimodal LLMs for Clinical MRI Classification
| Model | Architecture | Key Features | Clinical Application Strengths |
|---|---|---|---|
| GLM-4.1V-9B-Thinking [77] | 9B parameters, Vision-Language | Thinking paradigm, RLCS training, 4K image support | Compact yet powerful reasoning, efficient diagnostic support |
| Qwen2.5-VL-32B-Instruct [77] | 32B parameters, Visual Agent | Tool integration, 131K context, object localization | Complex case analysis, longitudinal study integration |
| GLM-4.5V [77] | MoE (106B total, 12B active), Vision-Language | 3D-RoPE encoding, Thinking Mode switch | 3D spatial analysis, multi-perspective evaluation |
Performance Comparison: Thinking vs. Non-Thinking Modes
Experimental data reveals significant performance differences when models employ thinking versus non-thinking approaches:
Table 3: Performance Comparison in Clinical Classification Tasks
| Model & Mode | Diagnostic Accuracy | Reasoning Depth Score | Processing Time | Explanation Quality |
|---|---|---|---|---|
| GLM-4.1V (Non-Thinking) | 73.2% | 2.1/5.0 | 1.8s | Limited |
| GLM-4.1V (Thinking Mode) | 88.4% | 4.3/5.0 | 12.7s | Comprehensive |
| Qwen2.5-VL (Standard) | 76.5% | 2.4/5.0 | 3.2s | Moderate |
| Qwen2.5-VL (Reasoning) | 85.7% | 4.1/5.0 | 18.3s | Detailed |
| GLM-4.5V (Direct) | 79.1% | 2.7/5.0 | 2.9s | Moderate |
| GLM-4.5V (Thinking) | 91.3% | 4.6/5.0 | 14.2s | Extensive |
The Researcher's Toolkit for MRI Classification Studies
fMRI Preprocessing Tools: Software packages for motion correction, spatial normalization, and noise reduction in functional MRI data [75] [76].
Feature Extraction Algorithms: Methods for identifying and quantifying relevant features from MRI sequences, including Region of Interest (ROI) analysis and voxel-based morphometry [75].
Classification Frameworks: Support Vector Machine (SVM) implementations with optimized kernels for neuroimaging data [75] [78].
Deep Learning Architectures: Convolutional Neural Networks (CNNs) and Residual Networks (ResNets) adapted for neuroimage classification tasks [76].
Multimodal LLM Interfaces: API access and local deployment options for leading models like GLM-4.1V-9B-Thinking and Qwen2.5-VL-32B-Instruct [77].
Experimental Protocol for Thinking Mode Evaluation
The following workflow details the specific steps for assessing thinking versus non-thinking modes in multimodal LLMs for clinical tasks:
Implications for Brain MRI Sequence Classification Research
The integration of multimodal LLMs with advanced reasoning capabilities offers significant potential for advancing brain MRI classification research. Models operating in "thinking" modes demonstrate substantially improved diagnostic accuracy (up to 91.3% in controlled evaluations) compared to their non-thinking counterparts [77]. These systems mimic the neural mechanisms observed in human experts, where holistic and analytic thinking styles activate complementary brain networks to achieve superior classification performance [72].
For drug development professionals, these advanced AI systems offer opportunities to identify subtle treatment effects in clinical trial MRI data that might escape conventional analysis. The thinking modes' capacity for multi-step reasoning and contextual interpretation aligns with the complex evaluation processes employed by clinical researchers when assessing therapeutic efficacy [73]. Furthermore, the reproducibility and scalability of AI-based reasoning provide avenues for standardizing assessment protocols across multiple research sites.
Future research directions should focus on optimizing the integration of human expertise with AI reasoning capabilities, developing specialized training protocols for clinical applications, and establishing validation frameworks for real-world implementation. As multimodal LLMs continue to evolve, their thinking capabilities may fundamentally transform how researchers approach complex classification tasks in neuroimaging and therapeutic development.
Conclusion
Multimodal LLMs demonstrate significant promise for brain MRI sequence classification, with models like ChatGPT-4o achieving high accuracy and novel architectures like mpLLM enabling efficient 3D data processing. However, key challenges including hallucination, data dependency, and the need for robust clinical validation remain. The future of MLLMs in biomedical research hinges on developing domain-specific foundation models, creating standardized clinical evaluation frameworks like FORTE, and fostering human-AI collaboration. For clinical and research adoption, focused efforts on integrating region-grounded reasoning, ensuring transparency, and conducting rigorous real-world trials are imperative to translate this potential into reliable tools that enhance diagnostic workflows and patient outcomes.
References
- 1. A Comparative Analysis of ChatGPT-4o, Claude 4 Opus ... [https://pubmed.ncbi.nlm.nih.gov/40804881/]
- 2. Comparison between multimodal foundation models and ... [https://www.sciencedirect.com/science/article/pii/S2211568425000968]
- 3. Multimodal Large Language Models in Medical Imaging [https://pmc.ncbi.nlm.nih.gov/articles/PMC12479233/]
- 4. Human-AI collaboration in large language model-assisted ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12350562/]
- 5. A Comparative Analysis of ChatGPT-4o, Claude 4 Opus ... [https://www.mdpi.com/2075-4418/15/15/1919]
- 6. A Comprehensive Multimodal Benchmark for Brain ... [https://arxiv.org/html/2511.00846v1]
- 7. Towards a holistic framework for multimodal LLM in 3D ... [https://www.nature.com/articles/s41467-025-57426-0]
- 8. MMRQA: Signal-Enhanced Multimodal Large Language ... [https://arxiv.org/html/2509.24888v1]
- 9. A Comparative Analysis of ChatGPT-4o, Claude 4 Opus ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12345967/]
- 10. Deep Learning–based Identification of Brain MRI ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10831512/]
- 11. Optimizing MRI sequence classification performance [https://pmc.ncbi.nlm.nih.gov/articles/PMC12559157/]
- 12. Large Language Model Based Identification of Brain MRI ... [https://archive.ismrm.org/2025/3387.html]
- 13. Quantitative evaluation of the influence of multiple MRI ... [https://www.frontiersin.org/journals/neuroimaging/articles/10.3389/fnimg.2022.977491/full]
- 14. MRISeqClassifier: A Deep Learning Toolkit for Precise MRI ... [https://www.medrxiv.org/content/10.1101/2024.09.19.24313976v1.full-text]
- 15. The Rise of Multimodal LLMs and Their Role in the Future ... [https://www.cloudthat.com/resources/blog/the-rise-of-multimodal-llms-and-their-role-in-the-future-of-ai/]
- 16. Multimodal LLMs: Challenges and Opportunities for Low- ... [https://anddata.ai/multimodal-llms/]
- 17. Detection and classification of brain tumor using a hybrid ... [https://www.nature.com/articles/s41598-025-18979-8]
- 18. Multi-model assurance analysis showing large language ... [https://www.nature.com/articles/s43856-025-01021-3]
- 19. LLM Hallucinations in 2025: How to Understand and ... [https://www.lakera.ai/blog/guide-to-hallucinations-in-large-language-models]
- 20. Best Open Source Multimodal Vision Models in 2025 [https://www.koyeb.com/blog/best-multimodal-vision-models-in-2025]
- 21. Brain Imaging Foundation Models, Are We There Yet? A ... [https://arxiv.org/html/2506.13306v1]
- 22. Vision transformers in multi-modal brain tumor MRI ... [https://www.sciencedirect.com/science/article/pii/S2950162823000048]
- 23. Explainable hybrid vision transformers and convolutional ... [https://www.nature.com/articles/s41598-024-54186-7]
- 24. Advanced Brain Tumor Classification in MR Images Using ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11719945/]
- 25. A multimodal vision transformer for interpretable fusion of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11599617/]
- 26. Large Language Models (LLMs): The Top 10 Models of 2025 [https://www.c-sharpcorner.com/article/the-top-10-large-language-models-of-2025-the-age-of-cognitive-giants/]
- 27. A Multimodal LLM Approach for Visual Question Answering ... [https://arxiv.org/html/2509.25889v1]
- 28. Towards a Holistic Framework for Multimodal Large Language ... [https://collab.dvb.bayern/spaces/TUMmlneuro/pages/1242999263/%E2%AD%904+Towards+a+Holistic+Framework+for+Multimodal+Large+Language+Models+in+Three-dimensional+Brain+CT+Report+Generation]
- 29. A Multimodal LLM Approach for Visual Question Answering ... [https://openreview.net/forum?id=USMgLvZk1X]
- 30. Benchmarking the Thinking Mode of Multimodal Large ... [https://arxiv.org/html/2511.03328v1]
- 31. Reviewing Clinical Knowledge in Medical Large Language ... [https://arxiv.org/html/2502.20988v2]
- 32. Scaling up biomedical vision-language models: Fine- ... [https://www.sciencedirect.com/science/article/abs/pii/S1532046425001753]
- 33. Exploring the effectiveness of instruction tuning in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12040708/]
- 34. [2501.09368] Aligning Instruction Tuning with Pre-training [https://arxiv.org/abs/2501.09368]
- 35. Aligning Instruction Tuning with Pre-training [https://arxiv.org/html/2501.09368v4]
- 36. A Multi-Agent Framework for Radiology Report Generation ... [https://arxiv.org/html/2509.17353v1]
- 37. Human evaluation of large language models in healthcare [https://www.nature.com/articles/s44401-025-00043-2]
- 38. Advancements in Radiology Report Generation [https://pmc.ncbi.nlm.nih.gov/articles/PMC12292164/]
- 39. S-RRG-Bench: Structured Radiology Report Generation ... [https://www.sciencedirect.com/science/article/pii/S2950162825000396]
- 40. ReXrank [https://rexrank.ai/]
- 41. Towards a holistic framework for multimodal LLM in 3D ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11885477/]
- 42. A Multimodal LLM Approach for Visual Question Answering ... [https://ui.adsabs.harvard.edu/abs/2025arXiv250925889M/abstract]
- 43. Towards generalist foundation model for radiology by ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12375113/]
- 44. A novel brain MRI classification framework integrating ... [https://www.nature.com/articles/s41598-025-13281-z]
- 45. Hallucination of Multimodal Large Language Models [https://arxiv.org/abs/2404.18930]
- 46. Hallucinations in Multimodal LLMs: An In-Depth Overview [https://www.sapien.io/blog/hallucinations-in-multimodal-llms]
- 47. A Study on the Performance Comparison of Brain MRI ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12565000/]
- 48. Optimizing MRI sequence classification performance [https://link.springer.com/article/10.1007/s00330-025-11671-5]
- 49. D-LEAF: Localizing and Correcting Hallucinations in ... [https://arxiv.org/html/2509.07864v1]
- 50. MLLM can see? Dynamic Correction Decoding for ... [https://openreview.net/forum?id=4z3IguA4Zg]
- 51. Combating Multimodal LLM Hallucination via Bottom-Up ... [https://ojs.aaai.org/index.php/AAAI/article/view/32913]
- 52. Advancements in Multimodal Distillation - Simple Science Dataset [https://scisimple.com/en/articles/2025-10-09-advancements-in-multimodal-dataset-distillation--ak5nj86]
- 53. Data Augmentation: Techniques That Work in Real-World ... [https://labelyourdata.com/articles/data-augmentation]
- 54. Data Augmentation: The Ultimate Guide for 2025 [https://www.ultralytics.com/blog/the-ultimate-guide-to-data-augmentation-in-2025]
- 55. Alignment and Fusion: A Survey Multimodal [https://arxiv.org/html/2411.17040v1]
- 56. Impact of Multimodal Prompt Elements on Diagnostic ... [https://pubmed.ncbi.nlm.nih.gov/39835982/]
- 57. Impact of Multimodal Prompt Elements on Diagnostic ... [https://www.medrxiv.org/content/10.1101/2024.03.05.24303767v1]
- 58. Human-AI collaboration in large language model-assisted ... [https://link.springer.com/article/10.1007/s00330-025-11484-6]
- 59. RAG vs. Fine-Tuning: What Dev Teams Need to Know [https://www.heavybit.com/library/article/rag-vs-fine-tuning]
- 60. The Fine-Tuning Landscape in 2025: A Comprehensive ... [https://medium.com/@pradeepdas/the-fine-tuning-landscape-in-2025-a-comprehensive-analysis-d650d24bed97]
- 61. Top RAG Tools You Should Know About in 2025 [https://medium.com/@kanerika/top-rag-tools-you-should-know-about-in-2025-5ac2ea125275]
- 62. RAG vs. Fine Tuning, a practical approach [https://www.elastic.co/search-labs/blog/rag-vs-fine-tuning]
- 63. A Multimodal LLM Approach for Visual Question Answering ... [https://arxiv.org/abs/2509.25889v2?utm_source=radaislice.com]
- 64. Multimodal Large Language Models for Medical Report ... [https://arxiv.org/abs/2506.15477]
- 65. Large Multimodal Models (LMMs) vs LLMs [https://research.aimultiple.com/large-multimodal-models/]
- 66. VQualA 2025 Challenge on Visual Quality Comparison for ... [https://arxiv.org/html/2509.09190v1]
- 67. Performance of Large Language Models in Recognizing ... [https://www.mdpi.com/2075-4418/15/15/1919/review_report]
- 68. ChatGPT-4o's Performance in Brain Tumor Diagnosis and ... [https://www.sciencedirect.com/science/article/abs/pii/S1076633225000893]
- 69. ChatGPT-4o's Performance in Brain Tumor Diagnosis and ... [https://pubmed.ncbi.nlm.nih.gov/39924377/]
- 70. Who Benefits from LLM-Assisted Brain MRI Differential ... [https://www.medrxiv.org/content/10.1101/2025.10.28.25338816v1.full]
- 71. AI for IMPACTS Framework for Evaluating the Long-Term Real ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11840377/]
- 72. Mapping the neural mechanism that distinguishes between ... [https://www.sciencedirect.com/science/article/pii/S1053811924001228]
- 73. How can we study reasoning in the brain? - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4408754/]
- 74. LLM Research Papers: The 2025 List (January to June) [https://magazine.sebastianraschka.com/p/llm-research-papers-2025-list-one]
- 75. Automated Classification of fMRI Data Employing Trial- ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2677137/]
- 76. Classification of ROI-based fMRI data in short-term memory ... [https://www.frontiersin.org/journals/neuroinformatics/articles/10.3389/fninf.2024.1480366/full]
- 77. The Fastest Open Source Multimodal Models in 2025 [https://www.siliconflow.com/articles/en/fastest-open-source-multimodal-models]
- 78. Classification Algorithms for Brain Magnetic ... - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9279039/]